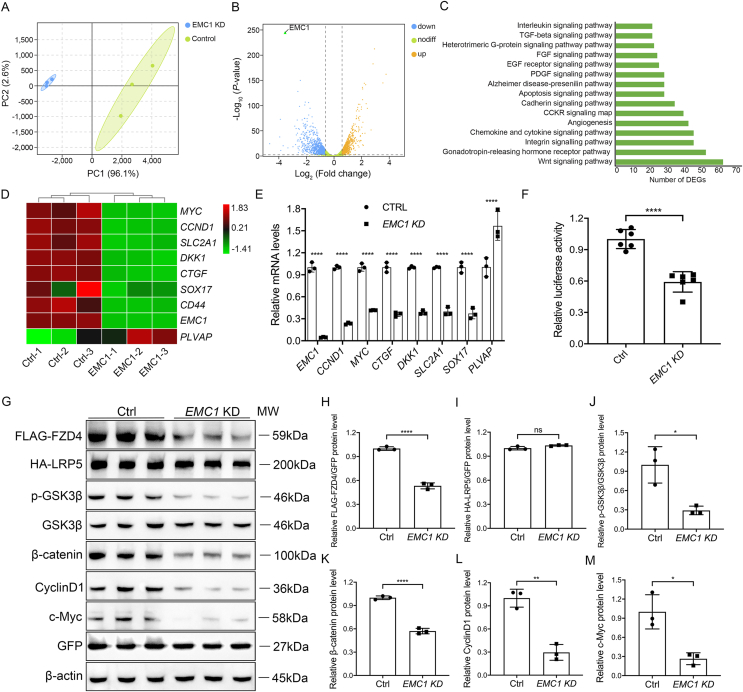
Figure 2.
EMC1 regulates Wnt signaling through FZD4 receptor. (A) Principal component analysis for Ctrl and EMC1 KD (knockdown) HRECs. (B) Volcano plot of differentially expressed genes (DEGs) in Ctrl and EMC1 KD HRECs. The dash lines indicate the threshold of P = 0.05 and |log2FC| = 0.585. The significantly down-regulated and up-regulated genes are shown as blue and yellow dots, respectively. Genes with no significant alterations are shown as green dots. (C) Pathway classification of DEGs using the PANTHER classification system online. (D) Heatmap of the expression fold changes of several Wnt-targeted genes in Ctrl and EMC1 KD HRECs as determined by RNA-seq analysis. (E) RT-qPCR quantification of the relative mRNA levels of EMC1, MYC, CCND1, SLC2A1, DKK1, CTGF, SOX17, CD44, and PLVAP in Ctrl and EMC1 KD HRECs. Error bars, SDs. Student's t-test (n = 3), ∗∗∗∗P < 0.0001. (F) Relative luciferase activity in Ctrl and EMC1 KD HEK 293STF cells transfected with pGL4-Renilla as an internal control. Error bars, SDs. Student's t-test (n = 6), ∗∗∗∗P < 0.0001. (G–M) Western blot and quantification analysis of the expression levels of FLAG-FZD4, HA-LRP5, p-GSK3β, β-catenin, CyclinD1, and c-Myc in Ctrl and EMC1 KD HEK 293T cells co-transfected with FLAG-tagged FZD4, HA-tagged LRP5, and GFP. GFP and β-actin were used as internal controls for over-expressed proteins and endogenous proteins (unless otherwise noted), respectively. GSK3β was used as an internal control for p-GSK3β (unless otherwise noted). Error bars, SDs. Student's t-test (n = 3), ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001. ns, not significant.