Abstract
Non-coding RNAs (ncRNAs) participate in the regulation of several cellular processes including transcription, RNA processing and genome rearrangement. The aberrant expression of ncRNAs is associated with several pathological conditions. In this review, we focused on recent information to elucidate the role of various regulatory ncRNAs i.e., micro RNAs (miRNAs), circular RNAs (circRNAs) and long-chain non-coding RNAs (lncRNAs), in metabolic diseases, e.g., obesity, diabetes mellitus (DM), cardiovascular diseases (CVD) and metabolic syndrome (MetS). The mechanisms by which ncRNAs participated in disease pathophysiology were also highlighted. miRNAs regulate the expression of genes at transcriptional and translational levels. circRNAs modulate the regulation of gene expression via miRNA sponging activity, interacting with RNA binding protein and polymerase II transcription regulation. lncRNAs regulate the expression of genes by acting as a protein decoy, miRNA sponging, miRNA host gene, binding to miRNA response elements (MRE) and the recruitment of transcriptional element or chromatin modifiers. We examined the role of ncRNAs in the disease pathogenesis and their potential role as molecular markers for diagnosis, prognosis and therapeutic targets. We showed the involvement of ncRNAs in the onset of obesity and its progression to MetS and CVD. miRNA-192, miRNA-122, and miRNA-221 were dysregulated in all these metabolic diseases. Other ncRNAs, implicated in at least three diseases include miRNA-15a, miRNA-26, miRNA-27a, miRNA-320, and miRNA-375. Dysregulation of ncRNAs increased the risk of development of DM and MetS and its progression to CVD in obese individuals. Hence, these molecules are potential targets to arrest or delay the progression of metabolic diseases.
Keywords: Biomarkers, CircRNAs, LncRNAs, Metabolic diseases, miRNAs, Non-coding RNAs, Therapeutic targets
Introduction
Non-coding RNAs (ncRNAs) are functional RNA molecules, transcribed from the genome but are not translated into proteins.1 Protein-coding RNAs represent only 3% of the human genome and ncRNAs account for approximately 97% of the total mammalian RNAs.2, 3, 4 ncRNAs of unknown function were previously considered as “biological dark matter or junk RNA”.5,6 Contrary to the established view, ncRNAs are now known to play a critical housekeeping and regulatory role in many cellular processes including transcriptional regulation, RNA processing, and genome rearrangement.7 miRNAs, circRNAs and lncRNA are major regulatory circulating ncRNAs and have participated in the regulation of gene expression and many cellular functions.8,9
Generally, an aberrant expression of ncRNAs is linked to the pathophysiology of many diseases and disorders.10,11 Advancements in high throughput sequencing techniques have unveiled the biomarkers and therapeutic potential of non-coding RNAs.10 In this review, we mainly focused on the role of regulatory circulating ncRNAs precisely, miRNAs, lncRNAs and circRNAs in metabolic diseases particularly, obesity, diabetes mellitus (DM), cardiovascular diseases (CVDs), and metabolic syndrome (MetS) pathogenesis and their importance as molecular biomarkers for diagnosis, prognosis and therapeutic targets of the diseases.
A systematic literature search on the clinical significance of ncRNAs in metabolic diseases was performed following PRISMA (preferred reporting items for systematic reviews and meta-analyses) (Fig. S1). Published articles related to the subject were identified using the search keywords: “clinical significance of ncRNAs in metabolic diseases”, “role of ncRNA in the pathogenesis of metabolic diseases”, “ncRNAs expression profile in metabolic diseases”, “role of ncRNAs in the pathogenesis of obesity, diabetes mellitus, cardiovascular diseases, and metabolic syndrome” etc. A total of 7793 articles were identified from Google Scholar, Embase, and PubMed central. Duplicates were removed and 4921 articles were left. A total of 3877 titles/abstracts were discarded beacause they were not focused on the area of the study. The remaining 1044 articles were evaluated for eligibility, in which only 59 articles were found to be eligible. The rest of the articles were discarded due to lack of relevant information, duplicates, or published earlier than 2011. Furthermore, 5 more relevant articles were discovered from manual searches and added to the list, which made a total of 64 unique, relevant, and full-text articles used for the synthesis of data.
Emphasis was given to explicate the role of regulatory circulating ncRNAs specifically miRNAs, circRNAs and lncRNAs in the pathogenesis and progression of the four most common metabolic diseases – obesity, DM, CVD, and MetS. The classification of ncRNAs is illustrated in Figure 1. Regulatory ncRNAs including miRNA, lncRNAs, and circRNAs are crucial regulators of gene expression affecting various cellular functions.12 Mechanistically, miRNA mediates gene regulation through several ways: 1) miRNAs interact with 3′ untranslated region (3′ UTR) of target genes with partial complementarity to induce translational repression or binds to 5′ UTRs to stabilize the target mRNAs and enhance the translation of target genes in animals13; 2) miRNA may bind to the target site within the coding exons with nearly exact matching to induce mRNA degradation particularly in plants13; 3) miRNAs are also reported to bind partially or impartially to the promoter region and decrease the activity of RNA Pol-II and recruit co-repressor or increase the stability of the pre-existing repressor complex, ultimately resulting in the transcriptional silencing of the target genes12; 4) Unconventionally, miRNAs may bind to the enhancer region and promote the transcriptional activation of genes through chromatin remodeling.12 However, circRNAs modulate gene regulation indirectly via their miRNA sponging activities, interacting with RNA binding protein and polymerase II transcription regulation.14 lncRNAs regulate target genes by acting as protein decoys, and/or miRNA sponging molecules. In addition, lncRNA binds to the miRNA response element (MRE) and masks the miRNA target site enhancing the stability and expression of the target gene. They are also reported to act as host genes to promote the miRNA generation (e.g., lnc-H19 acts as a reservoir of miRNA-675–5p and miRNA-675–3p).15,16 lncRNA also regulates gene expression by recruiting chromatin modifiers, or transcription factors to the promoter region of the target gene.16
Figure 1.
Classification of non-coding RNAs on the bases of functional characteristics and transcript size. ncRNAs can be classified into two major groups based on their functional characteristics: i) The housekeeping ncRNAs e.g., tRNAs, rRNAs, snoRNA, snRNAs and TER. ii) Regulatory ncRNAs. Regulatory ncRNAs are further classified on the basis of their transcripts size or number of nucleotides as either sncRNA or lncRNA. sncRNAs play a vital role in regulation of gene expression and include miRNAs, siRNAs, piRNAs. Other sncRNAs, e.g., tRNAs, rRNAs and snRNAs, are conventional RNAs that participate in the regulation of transcriptional and translational events. lncRNAs are further classified as intergenic lncRNAs, intronic lncRNAs, anti-sense lncRNAs, sense lncRNAs, elncRNAs, palncRNA and circular RNAs on the basis of genomic location. lncRNAs are also classified as cis- and trans-lncRNA based on the effect exerted on DNA. elncRNAs, enhancer associated lncRNA; lncRNAs, long non-coding RNAs; miRNA, microRNAs; ncRNAs, non-coding RNAs; palncRNAs, promoter-associated lncRNA; piRNA, P-element-induced wimpy testis (PIWI) interacting RNAs; rRNAs, ribosomal RNAs; siRNA, small interfering RNAs; snRNAs, small nuclear RNAs; sncRNA, short or small non-coding RNAs; snoRNAs, small nucleolar RNAs; tRNAs, transfer RNAs; TER, telomerase RNA.
Non-coding RNAs and metabolic diseases
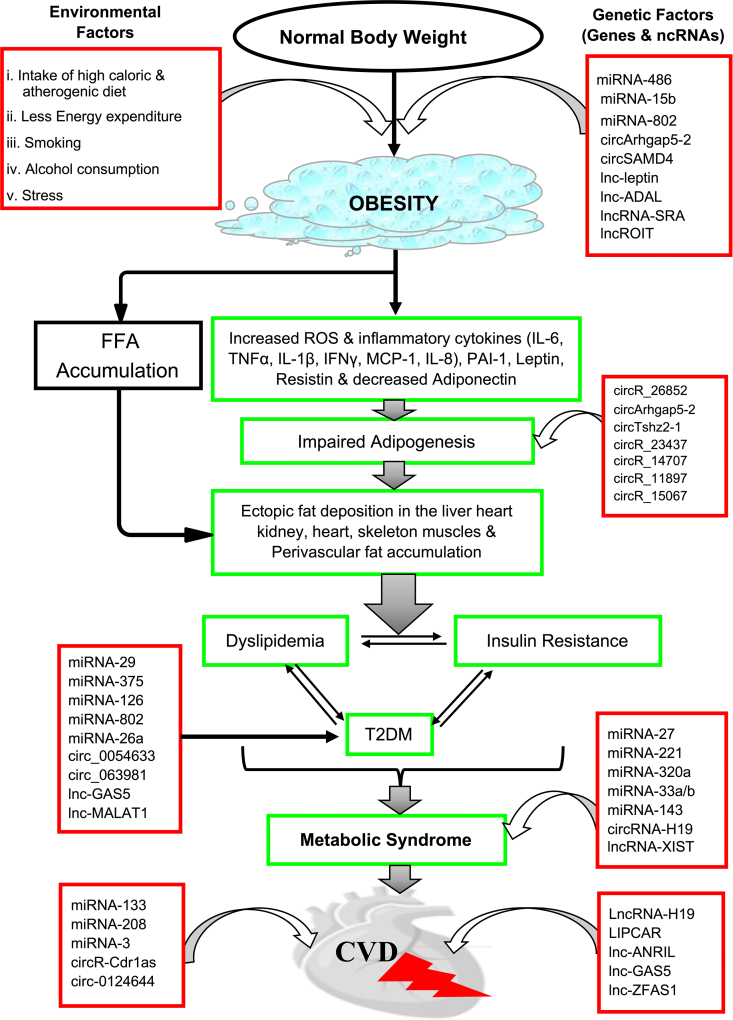
Non-coding RNAs play a critical role in the maintenance of metabolic homeostasis. Aberrant expression of ncRNAs is associated with the onset as well as progression of metabolic disorders. Thus, several ncRNAs have been identified as a potential molecular marker and/or therapeutic target of many metabolic diseases e.g., obesity, type 2 DM (T2DM), MetS, and CVD (Fig. 2; Fig. 3).
Figure 2.
Involvement of ncRNAs in the pathogenesis and advancement of metabolic diseases. Dysregulation of ncRNAs in a normal individual contributes to the onset of obesity and its progression to MetS and CVD. ncRNAs participate in various levels of the disease progression, and contribute to the impaired adipogenesis that causes the deposition of fatty substances on heart, liver, kidney and skeletal muscle as well as perivascular fat accumulation. Interlinked and central components of MetS (dyslipidemia, insulin resistance and T2DM) develop due to the fat accumulation on various organs. Large number of ncRNAs are involved and have contributed to the eventual setting of MetS and CVD. CVD, cardiovascular disease; FFA, free fatty acids; ROS, reactive oxygen species; T2DM, type 2 diabetes mellitus.
Figure 3.
The pattern of expression of ncRNAs dysregulates metabolic diseases. Many miRNAs are dysregulated in more than one metabolic disease, however, most of the circRNAs and lncRNAs are dysregulated in only one metabolic disorder. For example lnc-GAS5 is down-regulated in T2DM and up-regulated in CVD. miRNA-122 and miRNA-221 were down-regulated in all the metabolic diseases captured (obesity, T2DM, MetS and CVD). miRNA-192 was down regulated in CVD but up-regulated in obesity, T2DM and MetS. circ, circular RNA; CVD, cardiovascular diseases; lnc, long-chain non-coding RNA; MetS, metabolic syndrome; miRNA, microRNA; ncRNA, non-coding RNA; T2DM, type 2 diabetes mellitus.
Non-coding RNAs in obesity
Obesity is a heterogeneous metabolic disorder characterized by complex biogenesis, attributed to a sedentary lifestyle, overeating, and genetic predisposition.17 It is considered as the fundamental risk factor for the progression of diverse metabolic diseases including T2DM, hypertension, and CVD.18 Obesity and its related complications continue to rise in an unprecedented manner especially in developed countries and there is no approved drug for its cure. Dietary control and exercise are still the best way to manage obesity and its related metabolic complications. Besides other medical and physiological investigations, promising research on ncRNAs is underway to provide mechanistic insights into the role of ncRNAs in the onset and progression of obesity and other related metabolic diseases. The list of ncRNAs and their pattern of expression in obesity, as well as the biological processes affected, are presented in Table 1.
Table 1.
Summary of non-coding RNAs dysregulated in obesity and involved in its pathophysiology.
| Non-coding RNAs | Disease/Pathophysiology | Expression pattern | Cellular process | Subjects | Reference |
|---|---|---|---|---|---|
| miRNA-122 | obesity | up-regulated | glucose and lipid metabolism | mice | 24 |
| miRNA-192 | |||||
| miRNA-27a-3p | |||||
| miRNA-27b-3p | |||||
| miRNA-483-5p | obesity & IR | up-regulated | ? | human | 20 |
| miRNA-21 | obesity | down-regulated | ? | human | 21 |
| miRNA-138 | obesity &DM | down-regulated | ? | 19 | |
| miRNA-376a | |||||
| miRNA-15b | up-regulated | ? | 19 | ||
| miRNAe142-3p | obesity | up-regulated | human | 22 | |
| miRNA-140-5p | ? | ||||
| miRNA-143 | |||||
| miRNA 130 | |||||
| miRNA-532-5p | obesity | down-regulated | human | 22 | |
| miRNA-423-5p | ? | ||||
| miRNA-146 | |||||
| miRNA-520c-3p | |||||
| miRNA-15a | |||||
| miRNA-222 | obesity & T2DM | up-regulated | human | 23 | |
| miRNA-26b | ? | ||||
| miRNA-20a | |||||
| miRNA-146a | |||||
| miRNA-146b | |||||
| miRNA-15b | |||||
| miRNA-486 | obesity & T2DM | up-regulated | preadipocytes proliferation | human | 23 |
| miRNA-197 | obesity & T2DM | down-regulated | adipocyte differentiation & lipid metabolism | human | 23 |
| miRNA-802 | obesity | over expression | Insulin synthesis | mice | 26 |
| miRNA-122 | obesity | inhibition | chlesterol metabolism | mice | 46 |
| miRNA-26a | NAFLD | overexpression | ER Stress | mice | 31 |
| circRNA_26852 | adipogenesis | up-regulated | ? | pig | 34 |
| circRNA_11897 | adipogenesis | down-regulated | adipocyte differentiation & lipid metabolism | pig | 34 |
| circRNA_15067 | adipogenesis | down-regulated | pig | 34 | |
| circRNA_14707 | ? | ||||
| circRNA_23437 | |||||
| circ_0017650 | visceral obesity | up-regulated | fatty acids Metabolism | HPA-v & human | 36 |
| circ_0136134 | |||||
| circRNA9227-1 | |||||
| circArhgap5-2 | adipogenesis | up-regulated | adipogenesis | human& mice | 37 |
| circTshz2-1 | |||||
| circSAMD4A | obesity | up-regulated | preadipocyte differentiation | human & mice | 37 |
| lncRNA-p19461 | obesity | down-regulated | toll-like receptor (pro-inflammatory) pathway | human | 38 |
| lncRNA-p5549 | |||||
| lncRNA-p21015 | |||||
| ncRNA-SRA | obesity | overexpression | phosphorylation of Akt &FOXO1, | ST2 adipocytes | 39 |
| ncRNA-SRA | obesity | knock-down | adipocyte differentiation, phosphorylation of Akt & FOXO1 | 3T3-L1 preadipocytes | 39 |
| linc-ADAL | obesity | up-regulated | adipocyte differentiation & lipogenesis | human | 40 |
| lncR-ROIT | obesity | down | ubiquitin–proteasome pathway | mice | 44 |
| lncR-RP11-20G13.3 | childhood obesity | up-regulated | adipogenesis, inflammatory responses & lipid metabolism | human | 45 |
| lncRNA GYG2P1 | childhood obesity | down-regulated | ? | human | 45 |
miRNAs in obesity
Several reports have shown the involvement of miRNAs in obesity and its related complications. Pescador et al19 reported that miRNA-138, miRNA-15b, and miRNA-376a are predictive biomarkers of obesity. miRNA-138 and miRNA-376a were used as a powerful predictive tool for distinguishing obese patients from healthy individuals, diabetic subjects, and obese diabetic patients. It was further explained that miRNA-138 is implicated in the obesity by enteracting with 3′UTR of EID-1, which is potent inhibitor of adipogenic differentiation that interferes with SHP, an endogenous enhancer of adipogenic peroxisome proliferator-activated receptors (PPARγ2), a well-known transcription factor that regulates adipogenic gene expression.19 The association of miRNA-483-5p with obesity and IR, mediating the onset of DM, has been reported. miRNA-483-5p might have participated in the development of obesity and its related disorders, particularly insulin resitance, DM and CVD, through the regulation of insulin like growth factor 2 gene (IGF-2) and suppressor of cytokines signaling 3 gene (SOCS3).20 Ghorbani et al21 found that the expression level of miRNA-21 was negatively correlated with body mass index (BMI), waist circumference, insulin, and homeostasis assessment of IR (HOMA-IR) levels. Al-Rawaf22 highlighted the increased expression of miRNA142-3p, miRNA-140-5p, miRNA-143 and miRNA 130 in adolescents with obesity. On the other hand, miRNA-532-5p, miRNA-423-5p, miRNA-520c-3p, miRNA-146a, and miRNA-15a were significantly decreased in obese subjects. In addition, a strong association of these molecules with leptin and adiponectin was observed in the adolescent group with severe obesity. The up-regulated expression of miRNA-222, miRNA-486, miRNA-146b, miRNA-146a, miRNA-20a, miRNA-15b and miRNA-26b in obese subjects was also observed. The expression of miRNA-146b, miRNA-15b, miRNA-222, miRNA-15b and miRNA-486 was at least 6-fold higher in obese subjects as compared to non-obese. However, miRNA-197 expression was significantly down-regulated in obesity. Of these, miRNA-15, miRNA-146b and miRNA-486 are most important in forecasting the risk of developing DM in obese children.23 miRNA-486 was discovered to be involved in accelerating preadipocyte proliferation and myotube glucose intolerance, whereas miRNA-15, miRNA-146b, and miRNA-15b were discovered to be involved in suppressing high concentration glucose-induced pancreatic insulin secretion. These factors contributed to the pathological processes of obesity and T2DM. It was suggested that augmented expression of miRNA-122, miRNA-192, miRNA-27a-3p, and miRNA-27b-3p played a significant role in the initiation of metabolic syndrome (MetS); characterized by hyperglycemia, dyslipidemia, IR, and central obesity in the experimental mice.24 Dysregulation of miRNAs in adipocytes of obese subjects was observed. During adipogenesis, miRNAs can accentuate or halt adipocyte differentiation, thus regulate the development of fatty tissues.25 Notably, miRNA-27 and miRNA-519d target the PPAR family, which are crucial regulators of fat cell development.25 It was discovered that miRNA-802 plays a role in the development of obesity-related pancreatic β-cell dysfunction. The increased expression of miRNA-802 in high-fat diet (HFD)-induced obese mice is responsible for impaired insulin synthesis and secretion, resulting in impaired glycemic control. Mechanistically, miRNA-802 inhibits the expression of NeuroD1 and Fzd5, thereby regulating the phosphorylation of CREB, which suppresses the expression of Sox6. Furthermore, by regulating Fzd5, miRNA-802 increases calcium influx, resulting in reduced insulin secretion.26 miRNA-122 has been extensively studied as a serum biomarker for the severity of hepatocyte injury, which results in miR-122 release into the circulation. This miRNA was even found to be elevated prior to drug-induced liver damage.27 Non-alcoholic fatty liver disease (NAFLD) is one of the most serious complications of obesity.28,29 The disease is linked to the dysregulation of both protein coding genes and ncRNAs.28,30 Endoplasmic reticulum (ER) stress caused by NAFLD specifically induced the expression of miRNA-26a to counteract the stress on the liver cells and prevent lipid accumulation by controlling the expression of eukaryotic initiation factor 2 (eIF2). Down-regulation of miRNA-26a, on the other hand, aggravates this condition.31
circRNAs in obesity
Several circRNAs modulate the regulation of adipogenesis, which is well known biochemical event associated with the onset and development of obesity and its related complications.32 The role of circRNAs in adipogenesis signifies their role in the etiology of obesity and possible therapeutic benefit of the disease. Impairment of adipogenesis leads to abnormal fat deposition in the liver, kidney, and skeletal muscle, stimulating IR and increases the risk of its related disaeses.33 In a recent report, circRNA_26852 was significantly up-regulated, and circRNA_15067, circRNA_23437, circRNA_14707, circRNA_11897 were significantly down-regulated in subcutaneous adipose tissues (SCAT).34 Importantly, the functional analysis further revealed that circRNA_26852 and circRNA_11897 targeted a significant number of genes involved in adipocytes differentiation and lipids metabolism. It was further observed that circRNA_26852 targets miRNA-874 and miRNA-486, while circRNA_11897 targets miRNA-27a and miRNA-27b-3p, and regualate their expression via a phenomenon called competing endogenous RNAs (ceRNAs) mechanism. The decresed expression of miRNA-27 due to circRNA_11897 activity, results to the increased expression of target genes including PPARγ, hence the regulation of adipogenic differentiation and lipid metabolism.34 Another study reported the critical role of circArhgap5-2 and circTshz2-1 in adipogenesis via impacting the main adipogenic pathways including lipid catabolism and anabolism conserved in human adipocytes. Independent of miRNAs, circArhgap5-2 enhances core adipogenic processes including lipid metabolism and biosynthesis in human by acting as drivers global transcriptional programme.35 The induced expression of hsa_circ_0017650, hsa_circ_0136134, and hsa-circRNA9227-1 was reported in human preadipocytes-visceral (HPA-v), predicting their potential as diagnostic marker of the disease. hsa_circ_0136134 is implicated in HPA-v differentiation via regulating the expression of its parental genes (lipoprotein lipase (LPL)), which are critical molecule for adipocyte differentiation.36
circSAMD4A is involved in the regulation of adipocytes differentiation and its expression is associated with poor prognosis in obese patients. In vitro experiments indicated that circSAMD4A regulated adipocytes differentiation via its sponging properties to miRNA-138-5p, thus enhanced the expression of target gene (EZH2), an important regulator of adipocytes lipid metabolism and adipogenic differentiation.37
lncRNAs in obesity
The involvement of lncRNAs in the development of obesity and its related complications has been extensively documented (Table 1). Sun et al38 reported a significant decrease in the expression levels of lncRNA-p19461, lncRNA-p5549, and lncRNA-p21015 in individuals with obesity. Peroxisome proliferator-activated receptor gamma (PPARγ) is considered as a master of the transcriptional regulator of adipogenesis. Steroid receptor RNA activator (SRA), a lncRNA, is linked to obesity via the regulation of PPARγ and co-activates PPARγ-dependent reporter gene expression. The induction of SRA promotes the differentiation of ST2 mesenchymal precursor cells into adipocytes. On the other hand, SRA knockdown resulted in the inhibition of 3T3-L1 pre-adipocyte differentiation.39 A recent study demonstrated that the lncRNA is the most abundant in adipose tissue of obese patients and significantly induced during adipose differentiation. It was further explained that lnc-ADAL cooperates with insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) and heterogeneous nuclear ribonucleoprotein U (hnRNPU) to regulate the process of adipocyte differentiation and lipogenesis.40
One of the most prominent lncRNAs critically involved in maintaining obesity is Lnc-leptin (lnc-lep) which regulates the level of leptin. A super hormone responsible for regulating energy balance and body weight. It binds to a receptor, LepRb, in the hypothalamus to exert its action. Leptin serum level is elevated in obesity and increases with the increase in body fat.41 Both deficiencies of leptin and leptin resistance are considered as the major drivers of the pathogenesis of obesity.42 In vivo and in vitro studies revealed that lnc-lep regulates and maintains the leptin expression and its dysregulation is significantly associated with obesity.43 A recent genome-wide study revealed a total of 3203 dysregulated lncRNAs in HFD-induced obese mice. lncRNA 1810019D21Rik (ROIT) was down-regulated in the pancreatic islets of the obese mice and obese humans with T2DM, which was fairly reversed by the Hnf1b overexpression. Moreover, overexpression of ROIT resulted in improved insulin secretion and glucose homeostasis, establishing the link between obesity and beta-cell dysfunction via lncRNA ROIT.44
A microarray analysis conducted in childhood obesity identified a total of 1268 differentially expressed lncRNAs. Three up-regulated lncRNAs (RP11-20G13.3, LINC00968 and AC011891.5) and three down-regulated lncRNAs (GYG2P1, RP11-529H2.1 and OLMALINC) were validated by qRT-PCR analysis. Further analysis showed the involvement of these lncRNAs in lipid metabolism inflammatory response and osteoclast differentiation, among others. The up-regulated lncRNA, RP11-20G13.3, showed a positive correlation with waist circumference, waist-hip ratio, body mass index (BMI), low-density lipoprotein cholesterol, leptin and fasting insulin level. On the other hand, down-regulation of lncRNA GYG2P1 was negatively correlated with waist circumference, BMI, triglycerides (TG) and fasting insulin. Thus, lncRNA RP11-20G13.3 and lncRNA GYG2P1 play a critical role in the development of childhood obesity.45
Therapeutic potentials of ncRNAs in obesity
Many reports have shown the therapeutic potential of ncRNAs in various form of metabolic diseases. miRNA-122 was identified as an important molecule in the regulation of fatty acid and cholesterol metabolism, implying its potential as a therapeutic biomarker of obesity.46 Recently, significant down- regulation of miRNA-26a in hepatocytes of NAFLD patients, suggesting that increasing miRNA-26a expression could be a good strategy for treating NAFLD.31 It is one of the most serious complications of obesity. A recent analysis has elucidated the role of miRNA-122, miRNA-223, miRNA-21, miRNA-194/192, miRNA-155, and miRNA-29 as important regulators for liver physiological and pathological processes and therapeutic target of various forms of liver diseases. It was further explained that miRNA-21 promoted the advancement of NAFLD, however, miRNA-122 and miRNA-223 ameliorated the development of NAFLD.47 The increase in miRNA-223 in hepatocytes was detected due to preferential uptake of miRNA-223-enriched extracellular vesicles (EVs) derived from neutrophils. When EV-derived miRNA-223 was internalized by hepatocytes, it inhibited hepatic inflammatory and fibrogenic gene expression, thus played a beneficial role in mitigating the progression of NAFLD.48 We, therefore, suggest that designing a small molecule that enhances the expression of miRNA-223 can be a promising strategy for the treatment of NAFLD. The induced expression of hsa_circ_0017650, hsa_circ_0136134, and hsa-circRNA9227-1 was reported in human preadipocytes-visceral (HPA-v), predicting their potential as diagnostic and therapeutic target for visceral obesity.36 As highlighted above, circSAMD4A is an important molecule for development of obesity.37 Knockdown of circSAMD4A using small interfering RNAs (siRNAs) significantly reduced food intake, lowered body fat, reversed weight gain and improved energy expenditure, glucose tolerance, and insulin sensitivity in the HFD-induced obese mice. It clarifies the pathological role of circSAMD4A and its potential as a therapeutic target for the treatment of obesity.37
Considering the relevance of cellular events affected by dysregulation of ncRNA in obese individual, experimental and animal models were used to validate the results. Several reports on ncRNAs have been identified miRNA-486, miRNA-15b, circArhgap5-2, CircSAMD4, Lnc-leptin and LncRNA RP11-20G13.3, lnc-ADAL, ncRNA-SRA and lncROIT as appropriate ncRNAs to use in diagnosis, prognosis and therapeutics of obesity. These molecules have been deregulated in obese individuals and their knock-down or overexpression has resulted in altered lipid metabolism and adipogenesis. These are central events in the onset and progression of obesity.
Non-coding RNAs in diabetes mellitus
Diabetes mellitus is a metabolic disorder characterized by inadequate insulin secretion and/or failure of insulin action which deregulates the metabolism leading to hyperglycemia. Diabetes mellitus and its associated complications have been identified as a major global health challenge. According to Natarajan et al,49 T2DM remains the most common type of diabetes accounting for nearly 90% of the entire diabetes cases and usually characterized by IR, whereas type 1 diabetes mellitus (T1DM) accounts for the remaining 10% of diabetes cases. Both types of diabetes can eventually result in devastating microvascular complications (i.e., nephropathy, retinopathy and neuropathy) and macrovascular complications (i.e., CVD). These are life-threatening especially when left untreated or diagnosed at later stages.49 Various forms of ncRNAs (miRNAs, circRNAs and lncRNAs) have been identified for their potential as either molecular marker or therapeutic target of DM. Many research groups are interested in understanding the underline molecular mechanism of this disease, designing a diagnostic tool for early detection, or preventing its progression to more detrimental conditions. Significant number of dysregulated miRNAs in DM was reported in many studies (Table 2).
Table 2.
Summary of non-coding RNAs dysregulated in diabetes mellitus and involved in its pathophysiology.
| Non-coding RNAs | Type od DM | Expression pattern | Cellular process | Subjects | Reference |
|---|---|---|---|---|---|
| miRNA-150 | T2DM | up-regulated | insulin signaling & release | human | 50 |
| miRNA-30a-5p | |||||
| miRNA-15a | T2DM | down-regulated | insulin signaling & release | human | 50 |
| miRNA-375 | |||||
| miRNA-103 miRNA-28-3p miRNA-29a miRNA-9 |
Risk of T2DM | down-regulated | ? | human | 51 |
| miRNA-30a-5p miRNA-150 |
Risk of T2DM | up-regulated | ? | human | 51 |
| miRNA-9 miRNA-375 |
Pre-DM &T2DM | up-regulated | ? | human | 52 |
| miRNA-24 | T2DM with CHD | down-regulated | YKL-40 signaling pathway | human | 53 |
| miRNA-192 miRNA-194 |
T2DM | up-regulated | ? | human | 54 |
| miRNA-126 | T1DM &T2DM | down-regulated | vascular inflammation | human | 55 |
| miRNA-199a-3p miRNA-30 family miRNA-221 miRNA-26 family |
T2DM | up-regulated | angiogenic Pathways | human | 57 |
| miRNA-125b-5p miRNA-365a-3p |
T1DM | up-regulated | axon guidance, Rap1, focal adhesion, and neurotrophin signaling pathway | human | 59 |
| miRNA-5190 miRNA-770–5p |
T1DM | down-regulated | human | 59 | |
| miRNA-26a | up-regulation | cell differentiation | human/mice | 76 | |
| miRNA-26a | Obese/D2M | down-regulation | insulin signaling | mice | 77 |
| miRNA-26a | Obese/T2DM | up-regulation | β cell proliferation | mice | 78 |
| miRNA-802 | Obese/T2DM | up-regulation | glucose homeostasis | mice | 60 |
| miRNA-132 miRNA-222 miRNA-29a |
GDM | down-regulated | cholesterol & lipid metabolism | human | 61 |
| miRNA-142-3p miRNA-142-5p miRNA-144 miRNA-27a miRNA-342-3p |
T1DM, T2DM & GDM |
up-regulated | ? | human | 62 |
| miRNA-199a-5p miRNA-29b, miRNA-126 |
T1DM, T2DM & GDM |
down-regulated | ? | human | 62 |
| miRNA-99b miRNA-122 |
DKD | up-regulated | ? | human | 63 |
| miRNA-20a miRNA-486 |
DKD | down-regulated | oxidative stress, inflammation &apoptosis. | human | 63 |
| circ_0054633 | DM | up-regulated | cell proliferation, migration & angiogenesis | HUVEC & human | 64 |
| circ_0054633 | DM | up-regulated | cell cycle & mitotic cell cycle arrest | human | 66 |
| cirRNA_0054633 circRNA_063981 circRNA_103410 circRNA_102682 |
GDM | up-regulated | regulation of pancreatic β cell proliferation & glucose metabolism | human | 65 |
| circRNA_063981 circRNA_100750 circRNA_406918 circRNA_104387 circRNA_404457 circRNA_100192 circRNA_103410 |
DR | up-regulated | ? | human | 79 |
| circANKRD36 | T2DM | up-regulated | inflammation-associated pathways | human | 69 |
| circRNA_15698 | DN | up-regulated | ECM related protein synthesis | mice | 70 |
| lncRNA-MALAT1 | DN, DR, DC | up-regulated | inflammatory responses & apoptosis | HK2-cells, RF/6A cell & human | 74 |
| lncRNA-GAS5 | T2DM | down-regulated | ? | human | 71 |
| lncRNA-GAS5 | T2DM & DN | up-regulated | proliferation and fibrosis | human | 72 |
| lncMALAT1 | DR | down-regulated | pyroptotic cell death | human | 73 |
Pre-DM, pre-diabetes mellitus; T2DM, type 2 diabetes mellitus; T1DM, type1 diabetes mellitus; GDM, gestational diabetes mellitus; DKD, diabetic kidney disease; DR, diabetic retinopathy; DN, diabetic nephropathy; ECM, extracellular matrix.
miRNAs in diabetes mellitus
The role of miRNAs in the pathophysiology of diabetes mellitus was documented. Recently, a CORDIOPREV study conducted by Jiménez-Lucena et al50 revealed that plasma levels of miRNA-150, miRNA-30a-5p, miRNA-15a, and miRNA-375 were dysregulated three years before the onset of pre-diabetes. Thus, these molecules can be used to assess the risk of developing T2DM, which could help individuals at high risk, to take preventive measures, halt the disease initiation, progression, or delay its occurrence. Another CORDIOPREV study conducted on Spanish population placed on dietary intervention (Mediterranean diet and low-fat diets) in addition to conventional drugs for the treatment of coronary heart disease (CHD), demonstrated that individuals with lower expression of circulating miRNA-103, miRNA-28-3p, miRNA-29a, and miRNA-9 and high circulating levels of miRNA-30a-5p and miRNA-150 are at higher risk of developing T2DM.51 The study conducted on the Bahraini population who have not received any form of treatment, revealed the up-regulation of miRNA-9 and miRNA-375 in pre-diabetic subjects, and their expression was progressively increased in T2DM subjects; hence, associated with the development of T2DM. Further analysis revealed that the expression levels of miRNA-9 and miRNA-375 were positively correlated with glycemic status.52 The inconsistency in the pattern of expression of miRNA-9 between the studies of Jiménez-Lucena et al51 and Al-Muhtaresh & Al-Kafaji52 could be due to genetic differences between Spanish and Bahraini population. The dietary and conventional drug intervention used in Jiménez-Lucena et al study,51 could be another reason for the down-regulation of miRNA-9. The expression of miRNA-24 was significantly decreased in T2DM patients with CHD and negatively correlated with the YKL-40 gene known to be elevated in both type 1 and 2 DM as well as CVD. It was therefore recommended that circulating miRNA-24 is a potential molecule for both prognostic and diagnostic biomarker for T2DM with CHD.53 In another study, it was discovered that significantly higher serum levels of miRNA-192 and miRNA-194 in T2DM subjects compared to non-diabetic individuals, independent of fasting glucose or HbA1c.54 Overall, these studies are suggestive of the potential role of miRNAs as risk factors,50 biomarkers for disease progression,51 and diagnosis of T2DM.53 Diabetes mellitus is one of the major contributors to cardiovascular mortality usually via thromboembolic complications. The expression levels of endothelial-specific miRNA-126 were significantly decreased in both, individuals with DM and those at risk of developing the disease. The decreased expression of miRNA-126 is accompanied by an increase in vascular inflammation in diabetic patients, whereas increased expression of miRNA-126 resulted in decreased thrombogenicity.55 Similarly, Giannella et al56 reported a significantly higher level of miRNA-126-3p in normal subjects compared to pre-diabetic and diabetic individuals. The correlation analysis between miRNA-126-3p and the markers of endothelial dysfunction revealed a negative association of miRNA-126-3p with CD62E+ MPs and miRNA126-3p with plasma glucose, whereas the association was positive between the miRNA and plasma level of antioxidants. In another study, a total of 9 miRNAs were found differentially expressed in T2DM patients compared to healthy controls. It was observed that both pro-angiogenic miRNAs (miRNA-199a-3p and miRNA-30 family) and anti-angiogenic miRNAs (miRNA-221 and miRNA-26 family) were up-regulated and have a strong association with angiogenic genes.57 miRNAs were also implicated in the pathogenesis of DM via its effect on insulin signaling. According to Dooley et al,58 miRNA-29a is one of the most expressed miRNAs in both the liver and pancreas and is an important positive regulator of insulin secretion. Interestingly, miRNA-29a and miRNA-29c negatively regulated insulin signaling by phosphatidylinositol 3-kinase activity in the liver. Satake et al59 used a next-generation sequencing-based miRNA platform and measured the blood level of almost all known circulating miRNAs in T1DM patients and found that 54 miRNAs are associated with elevated glucose levels. Further analysis revealed that miRNA-365a-3p, miRNA-125b-5p, miRNA-5190, and miRNA-770-5p exhibit strong correlation with HbA1c levels. The dysregulations of these miRNAs implicated in the development of diabetes and its complications. The functions of miRNA-802 in glucose homeostasis and insulin sensitivity was highlighted. It was reported that both obese mice model and obese patients have increased expression of miRNA-802 in hepatic tissue. Increased expression of miRNA-802 in obese mice resulted in impaired glucose homeostasis and diminished insulin sensitivity via the inhibition of Hnf1b. On the other hand, decrease expression of miRNA-802 resulted in the increased expression of Hnf1b which in turn enhanced both glucose tolerance and insulin sensitivity.60 Dysregulation of various miRNAs in placental tissue leads to gestational diabetes mellitus (GDM). Significantly lower serum levels of miRNA-132, miRNA-222, and miRNA-29a were observed in the early second trimester of women with GDM. The knockdown of miRNA-29a significantly increased insulin induced gene 1 (Insig1) expression and eventually increased the expression of phosphoenolpyruvate carboxykinase 2 (PCK2), which may lead to the elevation of blood glucose level. miRNA-29a was identified as a negative regulator of glucose level and this study suggested miRNA-132, miRNA-29a and miRNA-222 as potential molecular candidates for predicting GDM.61 Another recent finding demonstrated that miRNA-483-5p was associated with obesity and insulin resistance as well as new-onset of DM and CVD.23 Collares et al62 performed a comparative analysis for miRNAs expression in T1DM, T2DM, and GDM patients. A total of 9 miRNAs (miRNA-126, miRNA-1307, miRNA-142-3p, miRNA-142-5p, miRNA-144, miRNA-199a-5p, miRNA-27a, miRNA-29b, and miRNA-342-3p) were found common among these three types of DM.
Diabetic kidney disease (DKD) remains the most prevalent complication of T2DM and a major cause of the end-stage renal disease (ESRD) in diabetic individuals. The clinical significance of several miRNAs in T2DM related DKD has been reported. Regmi et al63 found dysregulated serum expression levels of miRNA-99b, miRNA-122, miRNA-20a and miRNA-486 in DKD patients. It was significantly related to clinical parameters, e.g., albuminuria, estimated glomerular filtration rate (eGFR), blood glucose and lipid profiles. Furthermore, using receiver operating characteristic (ROC) curve analysis, the authors predicted higher diagnostic accuracy for miRNA-99b than miRNA-486-5p, miRNA-122-5p, and miRNA-20a in DKD with an area under the curve (AUC) of 0.895, 0.853, 0.80, and 0.697, respectively. Thus, the miRNAs could be a potential biomarkers panel for the diagnosis of DKD.
circRNAs in diabetes mellitus
circRNAs play a critical role in the pathogenesis of DM and its related complications (Table 2). Clinical significance of hsa_circ_0054633 is reported in various studies.64, 65, 66 The expression level of hsa_circ_0054633 was increased significantly in response to the elevated glucose level in DM. Conversely, the decreased expression of hsa_circ_0054633 promoted high glucose-induced endothelial cell dysfunction.64 Further analysis revealed that hsa_circ_0054633 inhibited miRNA-218 expression in human umbilical vein endothelial cells (HUVECs) which enhanced ROBO1 and HO-1 genes, suggesting that hsa_circRNA-0054633 has a protective effect against high glucose-induced endothelial cell dysfunction. Wu et al65 noted induction of hsa_cirRNA_0054633, hsa_circRNA_063981, hsa_circRNA_103410, and hsa_circRNA_102682 in GDM. In addition, hsa_circRNA_0054633 displayed a significant diagnostic value at different stages of pregnancy. These studies suggested the potential role of hsa_circRNA_0054633 in the pathogenesis of GDM and could be considered as a potential biomarker for the disease diagnosis.65 In another experiment, it was reported that increased expression of hsa_cirRNA_0054633 in the peripheral blood of T2DM subjects. Further analysis revealed that hsa_circ_0054633 is important in the regulation of vital cellular processes, e.g., cell cycle, cell cycle arrest, and energy metabolism. It is worthy to note that the cell cycle regulates β-cells proliferation, and that abnormal β-cells proliferation leads to inadequate synthesis and release of insulin, which is a fundamental feature for DM. It was therefore suggested that hsa_circ_0054633 participates in the development of DM by manipulating the cell cycle and metabolic processes. In addition, hsa_circ_0054633 exhibits significant diagnostic characteristics for both pre-diabetic and T2DM.66
Another circRNA CDR1-AS holds its significance via modulation of insulin secretion. The sponging activities of CDR1-AS affected the expression of miRNA-7 which is known to regulate the expression of the PAX6 gene, an important factor to regulate insulin secretion. The expression of CDR1-AS was inversely related to miRNA-7 expression leading to up-regulation of PAX6 and eventually increased insulin secretion.67 In a recent study, at least 2600 circRNAs were identified in human islets cells in which circCIRBP, circZKSCAN, circRPH3AL, and circCAMSAP1 demonstrated significant associations with DM. Further analysis demonstrated a strong association of circCIRBP with insulin secretory index in isolated human islets; this provides important information about the potential role of circRNAs in the diagnosis of DM.68
A significantly higher expression of circANKRD36 was found positively correlated with IL-6, glucose and glycosylated hemoglobin in T2DM patients (r = 0.393, P = 0.031, r = 0.3250, P = 0.0047 and r = 0.3171, P = 0.0056, respectively). Further analysis revealed that circANKRD36 could interact with hsa-miRNA-498, hsa-miRNA-3614-3p, and hsa-miRNA-501-5p and interfere with T2DM and inflammation-associated pathways. Therefore, circANKRD36 could serve as a potential diagnostic marker for chronic inflammation in patients with T2DM.69 However, in vivo studies are required for verification of these findings. The up-regulation of circRNA_15698 was also observed in DR mice. A computational and luciferase reporter assay revealed that circRNA_15698 positively regulates the expression of transforming growth factor-β1 (TGF-β1) via its sponging activity on miR-185. Further analysis validated the role of circRNA_15698/miR-185/TGF-β1 in the biosynthesis of extracellular matrix (ECM)-associated protein.70
lncRNAs in diabetes mellitus
Dysregulation of lncRNAs is linked to both onset and progression of DM. lncRNAs play a pivotal role in many biological processes involved in the progression of DM and its related complications involving pancreatic beta-cell dysfunction, IR, and epigenetic regulation.12 Studies report the association of lncRNA Gas5 with diabetes,71,72 and lncRNA-MALAT1 with diabetic nephropathy.68 A significantly decreased expression of lncRNA Gas5 was observed in the serum of diabetic patients as compared to non-diabetic individuals.71 In another study, in vivo data revealed that lncRNA GAS5 inhibited cell proliferation and fibrosis in diabetic nephropathy via its sponging effect on miRNA-221, which regulated the expression of SIRT1. In addition, circulating lncRNA-GAS5 was associated with the prevalence of T2DM.72
In diabetic nephropathy, lncRNA-MALAT1 down-regulation in HK2 cells (immortalized renal proximal tubule epithelial cells) inhibiteded hyperglycemia-induced pyroptosis via the up-regulation of miRNA-23c. An inhibition of MALAT1 resulted in the down-regulation of genes related to DM including Caspase-1, NLRP3, ELAVL1, and IL-1β. Surprisingly, similar result was obtained upon induction of miRNA-23c. Further analysis revealed that miRNA-23c is a target of lncRNA-MALAT1 and has repressed the expression of ELAVL1 leading to decreased expression of its downstream protein (NLRP3). The expression of MALAT1 prevented the negative effect of miRNA-23c on its target ELAVL1 thereby inhibiting hyperglycemia-induced cell pyroptosis.73 Dysregulation of lnc-MALAT1 was observed in many diabetic-related complications, i.e., ischemic reperfusion injury, retinopathy, cataract, atherosclerosis, cardiomyopathy, non-alcoholic steatohepatitis, gastroparesis, kidney disease, and GDM.74
Therapeutic potentials of ncRNAs in diabetes mellitus
Many studies have shown that ncRNAs are good candidates for the detection and monitoring the progress of DM. An in vivo study conducted by Jo et al75 revealed that miRNA-204 knockout in mice model promoted islet glucagon-like peptide 1 receptor (GLP1R) expression and improved sensitivity to GLP1R agonist. It resulted in improved glucose tolerance, insulin secretion and cAMP level; therefore, it protects the development of diabetes. Fu et al76 reported that miRNA-26a is necessary in pancreatic cells development via the regulation of ten eleven translocation (TET) enzymes. Both in vivo and in vitro experiment showed regulation of TET by the miRNA-26a in enhanced pancreatic cell differentiation. Additionally, the regulatory role of miRNA-26a on insulin signaling, glucose and lipid metabolism was ascertained. In mice fed a high-fat diet, overexpression of miRNA-26a enhanced insulin sensitivity, lowered hepatic glucose production, and reduced fatty acid synthesis, mitigating obesity-related metabolic complications. In contrast, suppressing endogenous miRNA-26a reduced insulin sensitivity, increased fatty acid and glucose synthesis. miRNA-26a targeted several key regulators of hepatic metabolism and insulin signaling. In similar reports, the significance of miRNA-26a in insulin sensitivity was demonstrated. miRNA-26a knockout in β cells enhanced obesity-induced metabolic disorders. Conversely, overexpressing of miRNA-26a in β-cells inhibited obesity-induced metabolic abnormalities with decreased insulin resistance and hyperinsulinemia occurred due to obesity. Thus, miRNA-26a can be a promising candidate for the treatment of T2D.77,78
Another study revealed the significant induction of hsa_circRNA_063981, hsa_circRNA_100750, hsa_circRNA_406918, hsa_circRNA_104387, hsa_circRNA_404457, hsa_circRNA_100192 and hsa_circRNA_103410 in the serum of patients with diabetic retinopathy (DR). Further analysis revealed that hsa_circRNA_104387 can potentially regulate the expression of miRNA-29a, however, circRNA_103410 can bind to the miRNA-126 and sequester its effect on the target genes (VEGF and MMP-9) in DR, results in endothelial injury and progression of vascular complications. This is evident that hsa_circRNA_104387 and circRNA_103410 play a vital role in the pathogenesis of DR by sequestering the effect of miRNA-29a and miRNA-126 rspectively, thus, can serve as a novel target candidate for treatment of the diabetes.79
As highlighted above, scientific investigations have confirmed the involvement of miRNA-29,51,61,75 miRNA-375, miRNA-126,55,56,62 miRNA-26a,76, 77, 78 hsa_circ_0054633,64, 65, 66 hsa_circ_063981,65,80 lnc-GAS5,71,72 and lncMALAT173,74 in DM and its complications. Therefore, further validation of these molecules has been emphasized, both as diagnostic marker as well as therapeutic target of diabetes mellitus.
The most effective therapeutic intervation for diabetes should control the hyperglycemia by regulating the synthesis and secretion of insulin. As well as normalizing cellular uptake of glucose by alleviating insulin resistance. These are the genesis of virtually all diabetes mellitus related complications. Therefore, insulin sensitivity and normal functions of pancreatic β-cells, are responsible for the synthesis, storage and secretion of insulin.81 The role of miRNA-26a in insulin sentivity and pancreatic β-cells development and differentiation is reported. Thus, a pharmacological agent that characteristically designed to boost the expression of miRNA-26a can improve the health status of diabetic patients. Recently, it was indicated that administration of metformin (antidabetic drug) results in a significant down-regulation of miRNA-26b in the brains of diabetics.82 This can be a mechanistic way by which metformin exhibits its antidiabetic effect in human. Thus, a molecule with a strong binding efficiency to miRNA-26b may act as potent antidiabetic agents.
Non-coding RNAs in cardiovascular diseases
Cardiovascular disease is a general name given to various conditions affecting the heart and blood vessels. The disease remains the major cause of death resulted from metabolic disorders. It claims the life of 17.9 million people annually and contributed to about 31% of the total death worldwide.83 Obesity related low-grade systemic inflammation, diabetes and hypertension are recognized as the major risk factors responsible for the onset and advancement of CVD to detrimental endpoint.84 Identification of the molecular events associated with the onset and progression of this disease would pave ways for the early diagnosis and treatment of CVD, which could reduce the detrimental effect of the disease. Many studies have unveiled the role of ncRNAs in cardiovascular events (Table 3).
Table 3.
Summary of non-coding RNAs dysregulated in cardiovascular diseases and involved in its pathophysiology.
| Non-coding RNAs | Type of CVD | Expression Pattern | Cellular process | Subject | Reference |
|---|---|---|---|---|---|
| miRNA-1 | AMI | up-regulated | human | 88 | |
| miRNA-133a | ? | ||||
| miRNA-133b | |||||
| miRNA-208 | AMI | up-regulated | human | 88 | |
| miRNA-320b | ? | ||||
| miRNA-499 | |||||
| miRNA-140-3p | CHD | up-regulated | hypoxia pathway | human | 91 |
| miRNA-132 | |||||
| miRNA-210 | |||||
| miRNA-33 | ATHC | up-regulated | mice | 86 | |
| miRNA-148a | |||||
| miRNA-128-1 | |||||
| miRNA-26 | ? | ||||
| miRNA-33 | |||||
| miRNA-106 | |||||
| miRNA-144 | |||||
| miRNA-758 | |||||
| miRNA-221 | CHD | up-regulated | ? | human | 105 |
| miRNA-222 | |||||
| miRNA-146a | |||||
| miRNA-146b | |||||
| miRNA-133 | CHD | down-regulated | ? | human | 92 |
| miRNA-208a | |||||
| miRNA-17 | |||||
| miRNA-92a | |||||
| miR-126 | |||||
| miR-145 | |||||
| miR-155 | |||||
| miRNA-122 | CHD | up-regulated | lipid metabolism | human | 93 |
| miRNA-126 | CHD | up-regulated | human | 94 | |
| miRNA-199a | |||||
| miRNA-375 | |||||
| miRNA-223 | AMI | down-regulated | ? | mice | 89 |
| miRNA-145 | |||||
| miRNA-125b | |||||
| miRNA-125a | |||||
| let-7b | |||||
| miRNA-208 | CHD | up-regulated | ? | human | 95 |
| miRNA-215 | |||||
| miRNA-487a | |||||
| miRNA-502 | |||||
| miRNA-29b | CHD | down-regulated | inflammation & fibrosis. | human | 95 |
| circ_0082081 | CHD | up-regulated | cell apoptosis & robo receptor signaling pathway |
human | 99 |
| circ_0098964 | |||||
| circ_0113854 | |||||
| circ_0124644 | |||||
| circRNA5974-1 | |||||
| ZFAS1 | AMI | up-regulated | cardiomyocytes apoptosis | mice | 100 |
| GAS5 | ATHC | up-regulated | lipids influx | mice | 107 |
| ANRIL | ATHC | down-regulated | Inflammatory response & cholesterol efflux | mice | 102 |
| miRNA-302a | ATHC | up-regulated | cholesterol homeostasis | mice | 106 |
| LIPCAR | CHD | up-regulated | oxidative phosphorylation & inflammasome activation | human | 80 |
| H19 | CHD | up regulatted | ? | 80 |
ATHC, atherosclerosis; CHD, coronary heart disease; CVD, cardiovascular diseases; MI, myocardial infarction.
miRNAs in cardiovascular diseases
The aberrant expression of miRNAs has been observed in stroke, coronary artery disease (CAD), peripheral vascular disease, and other vascular pathologies.85 The involvement of miRNAs in CVD-related cellular processes, e.g., inflammation, cholesterol homeostasis, oxidative stress, and hypertension, has been reported.85 A recently published study highlighted that miRNA-21, miRNA-155, miRNA-126, miRNA-146a, miRNA-146b, miRNA-143, miRNA-145, miRNA-223, and miRNA-221 are the most widely reported miRNAs in hypertension and atherosclerosis.86 The most prominent one is miRNA-21, due to its abundance in the blood vessel wall and the ability to respond to the shear or mechanical stress to the vessels.71 An elevated level of plasma miRNA-142 is significantly associated with major adverse cardiovascular events (MACE) among patients with coronary artery disease who had a percutaneous coronary intervention (PCI) and considered to be an independent risk factor for MACE. Thus, miRNA-142 could serve as a potential biomarker for MACE among CAD patients following PCI.87
The role of cardiomyocytes enriched miRNAs in the diagnosis of CVDs has been proposed earlier. The up-regulation of miRNA-1, miRNA-133a, miRNA-133b, miRNA-208 and miRNA-499 were reported in the plasma of acute myocardial infarction (AMI).88 On the other hand, miRNA-126, Let-7b, miRNA-125a, miRNA-125b, miRNA-145, miRNA-320b, miRNA-223, and miRNA-375 were down-regulated in AMI. This indicates the potential role of these miRNAs as molecular markers of the disease.89 In addition, a meta-analysis revealed that miRNA-133a, miRNA-133b, miRNA-499, miRNA-208a, and miRNA-208b can be potential candidates for diagnostic and prognostic of CVD.90 A study that investigated the role of miRNAs in cardiovascular mortality revealed that miRNA-140-3p, miRNA-132, and miRNA-210 are important predictors of death in individuals with CAD.91
Up-regulation of miRNA-221, miRNA-222, miRNA-146a, miRNA-146b, miRNA-122 and down-regulation of miRNA-133, miRNA-208a, miRNA-17, miRNA-92a, miRNA-126, miRNA-145, miRNA-155 were reported in CHD patients compared to healthy control group.92, 93, 94 Furthermore, the up-regulation of miRNA-126 and miRNA-199a in circulating micro-vesicles is associated with the decreased risk of cardiovascular events in stable CHD patients. This suggests that the expression of potential cardio-protective miRNAs in circulating micro-vesicles holds prognostic value in patients with stable CHD.94 It was observed that the expression levels of miRNA-208, miRNA-215, miRNA-487a, and miRNA-502 were significantly up-regulated, while miRNA-29b expression was down-regulated in the Chinese population with atypical coronary artery disease (ACAD) compared to healthy individuals. Furthermore, the ROC analysis revealed that the miRNA-208, miRNA-215, miRNA-487a, miRNA-502, and miRNA-29b have high diagnostic values for the detection ACAD. Additionally, computational analysis of these miRNAs predicted their involvement in the pathogenesis of CHD via the regulation of certain genes crucial in cardiac and vascular remodeling, especially in pathways of inflammation and fibrosis. Thus, the potential role of these miRNAs as therapeutic targets is suggested in CHD with an atypical presentation.95
circRNAs in cardiovascular diseases
The underline relationship between circRNAs and CVD has been elucidated in many studies.96, 97, 98, 99 The tissue-specific and the conservative nature of circRNAs and their regulatory roles make them attractive candidates for therapeutic targets. Recently, Jakobi et al96 have identified 63 conserved circRNAs expressed in human cardiovascular cell models (hiPSC-CMs and HUVECs) as well human cardiac tissue. Overexpression of circRNA CDR1-AS intensified myocardial infarction (MI) by acting as miRNA-7a sponge, which regulates the expression of poly ADP ribose polymerase (PARP), and SP1 genes known to play a pro-apoptotic role during the development ofMI.98 Significant up-regulation of hsa_circ_0082081, hsa_circ_0098964, hsa_circ_0113854, hsa_circ_0124644, and hsa-circRNA5974-1 was observed in CHD patients when compared to the control. Further analysis revealead that hsa_circ_0124644 has been linked to many cellular processes including cell apoptosis, and Robo receptor signalling pathway. These are vital proceses for the onset and development of CHD. Additionally, ROC curve analysis revealed that hsa_circ_0124644 has the highest sensitivity and specificity.99
lncRNAs and cardiovascular diseases
There is increasing evidence for the role of lncRNAs in cardiac-related illnesses. The regulatory role of a lncRNA-ZFAS1 is explored in AMI. The significance of lncRNA-ZFAS1 has been observed in the myocardium infarcted zone and the border zone of the AMI rats model where the expression of miRNA-150 was significantly decreased. As highlighted above, miRNA-150 has a protective effect against AMI. ZFAS1 knockdown or miRNA-150 overexpression showed a relieving effect on the MI rats. In addition, lncRNA-ZFAS1 possessed a potential binding site for miRNA-150 and can directly influence the expression of miRNA-150.100
As described above, atherosclerosis is the main underlying pathology of CVDs that develops over the years. Several reports highlighted the involvement of lncRNAs in several atherosclerotic processes, including endothelial dysfunction, inflammation and lipid deposition.69,80,90,92, 93, 94 Moreover, lncRNAs are expressed in many cell types present in atherosclerotic lesions.101
lncRNA CDKN2B-AS1 (cyclin-dependent kinase inhibitor 2B antisense 1), also called ANRIL, is predominately expressed in cardiac cells, vascular endothelial cells, and monocyte-derived macrophages.102 It was observed that many ncRNAs were dysregulated in CVD, and a few of them were further investigated for their potential role as therapeutic targets or diagnostic biomarkers in different types of CVD. The miRNA-3, lnc-ANRIL, and lnc-GAS5 genes can be used to treat atherosclerosis, and circRNA Cdr1as and lnc-ZFAS1 genes to treat MI. Based on these studies hsa-circ-0124644, lncRNA-H19, and LIPCAR may act as strong candidates for the diagnosis of CAD. Dysregulation of miRNA-133,88,90,92 and miRNA-20889,92,95 was reported recurrently, thus, could be further studied to validate their novelty as a diagnostic, prognostic and theraprutic target of CVD. Intake of flavonoids is one of the recommended intervation for the management of various form of CVDs, owing to its effects on the prevention of endothelial dysfunction, improvement of lipid metabolism, reduced oxidative stress and lowered blood pressure. Flavonoid confers a therapeutic benefit against CVDs via interaction with lncRNAs, thus a potential partnership between the lncRNAs and flavonoids in treating CVDs exist.84
Therapeutic potentials of ncRNAs in cardiovascular diseases
It has been reported that miRNA-150 acts as a potential regulator of monocyte migration and the production of pro-inflammatory cytokines, resulting in cardioprotective effects against AMI-induced injury and can be a potential candidate for therapeutic intervention in MI.103 Atherosclerosis is the fundamental cause of MI, peripheral artery disease, and ischemic stroke.104 The accumulation of low-density lipoprotein (LDL) leads to the activation of endothelial cells, which initiates inflammatory responses and recruitment of monocytes that eventually differentiate into macrophages, responsible for the absorption of modified lipoprotein and become foam cells.104 miRNA-30c inhibited the biosynthesis of lipids by acting on lysophosphatidyl glycerol acyltransferase-1 (LPGAT1), therefore an increased expression of miRNA-30c can be used in the treatment of hyperlipidemia and atherosclerosis.83 Evidences revealed that miRNA-33 knock-down resulted in increased plasma HDL levels and decreased atherosclerotic plaque size.105 ATP-binding cassette transporter (ABCA1) gene is a major player in controlling cholesterol efflux across the cell membrane onto lipid-poor apoA1 to mediate both hepatic HDL synthesis and the elimination of excess cholesterol from peripheral cells.105 It was discovered that miRNA-302a regulates ABCA1 expression and the inhibition of miRNA-302a resulted in decreased atherosclerosis development in the mice model.106 Similarly, inhibition of miRNA-148a, miRNA-128-1, miRNA-26, miRNA-33, miRNA-106, miRNA-144, and miRNA-758 resulted in the decrease of atherosclerosis build-up via the target of ABCA1.105 Thus, these miRNAs are suggested as potential targets to inhibit the development of atherosclerosis. In another study, circular antisense non-coding RNA in the INK4 locus (circANRIL) was identified as a potential target for the treatment of atherosclerosis. circANRIL confered an atheroprotective effect by regulating ribosomal RNA (rRNA) maturation and atherogenic pathways. Mechanistically, circANRIL binds to the C-terminal lysine-rich domain of pescadillo homologue 1 (PES1), thereby impairing exonuclease-mediated pre-rRNA processing, ribosome biogenesis in vascular smooth muscle cells and macrophages. This resulted in the activation of p53, thereby inducing apoptosis and inhibition of proliferation, which are fundamental cell functions in atherosclerosis. Thus, circANRIL can be considered as a promising target for the prevention and treatment of atherosclerosis.97
It was observed that the expression of ABCA1 can be inhibited by GAS5 by interacting with the enhancer of zeste homolog 2 (EZH2). EZH2 induction decreased both cholesterol efflux and ABCA1 expression. EZH2 can bind to the promoter region of ABCA1 and regulate its expression. lncRNA-GAS5 knock-down potentially enhanced the reverse-transportation of cholesterol, inhibited the accumulation of intracellular lipids and prevented the development of atherosclerosis via reducing EZH2-mediated transcriptional inhibition of ABCA1. The study suggested that lncRNA-GAS5 can be used as a therapeutic target for the management of atherosclerosis and its advancement to CAD.107 The down-regulation of ANRIL and significantly increased expression of A disintegrin and metalloprotease 10 (ADAM10) were observed in atherosclerotic plaque tissue and THP-1 macrophage-derived foam cells. Moreover, overexpression of ANRIL and knock-down of ADAM10 resulted in increased cholesterol efflux, and suppressed lipid accumulation in atherosclerotic plaque tissue and THP-1 macrophage-derived foam cells. Nuclear localized ANRIL recruited DNA methyltransferase 1 (DNMT1) and enhanced the methylation of ADAM10 promoter, accentuated cholesterol efflux and inhibited atherosclerotic inflammation.102 Therefore, a molecular strategy designed to concurrently increase the expression of ANRIL and decrease the expression of ADAM10 could have therapeutic benefit on atherosclerosis.
Coronary heart disease is one of the major forms of CVD and is primarily caused by atherosclerosis that decreases the blood flow to the heart muscles via coronary arteries. Dysregulation of miRNAs was observed in CAD patients and the miRNAs were recommended as a potential therapeutic target or molecular markers of the disease. In a recent report, Pan108 described a significant increase of lncRNA-H19 in atherosclerotic individuals as compared to the healthy control group. Similarly, overexpression of lncRNA-H19 in human umbilical vein endothelial cells (HUVEC) resulted in increased proliferation and decreased apoptotic death. Interestingly, p38 and p65 were also up-regulated due to lncRNA-H19 overexpression.108 In another report, the blood levels of lncRNA-H19 and LIPCAR were significantly increased in both CAD patients and patients with heart failure compared to an individual with normal cardiac function.80 Therefore, the consideration of H19 and long intergenic noncoding RNA predicting cardiac remodeling and survival (LIPCAR) as new molecular markers for CAD has been suggested. Cardiomyocyte senescence is a significant event that contributes to the deterioration of cardiac physiological function as well as the risk of developing CVD. H19 overexpression accelerated cardiomyocyte senescence and its knockdown inhibited the process, demonstrating its involvement in the senescence. H19 participates in this process by inhibiting the regulatory effect of miRNA-19a in a ceRNA mechanism, thereby regulating the expression of suppressor of cytokine signalling 1 (SOCS1) and activating the p53/p21 signalling pathway.109 All together, H19 is a potential target candidate for the treatment of CVD.
Non-coding RNAs in metabolic syndrome
Metabolic syndrome (MetS) is one of the major health challenges worldwide with an estimated case of 11.9%–37.1% in the Asian Pacific region, 11.6%–26.3% of the European population, and 12.5%–62.5% of the African population.110 MetS cases continue to increase with the rise of obesity and are at high risk of cardiovascular morbidity and mortality. It has been established that various components of MetS, i.e., obesity, dyslipidemia and T2DM, alter the expression of non-coding RNAs. On the other hand, dysregulation of ncRNAs mediates the onset and progression of many metabolically related disorders defining metabolic syndrome. Some of the important ncRNA implicated in the metabolic syndrome are presented (Table 4).
Table 4.
Summary of non-coding RNAs dysregulated in metabolic syndrome and involved in its pathophysiology.
| Non-coding RNAs | MetS | Pattern of Expression | Cellular process | Subject | References |
|---|---|---|---|---|---|
| miRNA-122 | MetS & T2DM | up-regulated | hepatic lipid metabolism | human | 111 |
| miRNA-196 miRNA-27b miRNA-301 |
MetS | up-regulated | P16/MAPK Pathway | MetS-MSCs | 112 |
| miRNA-320a miRNA-97-3p miRNA-23-3p miRNA-221-3p miRNA-27a-3p miRNA-130a-3p |
MetS & Obesity | down-regulated | insulin secretion, lipid and carbohydrate metabolism | human | 113 |
| miRNA-150 miRNA-192 miRNA-27a miRNA-320a |
MetS & T2DM | up-regulated | vascular signaling pathways, glucose & cholesterol metabolism | human | 114 |
| miRNA-27a miRNA-130b |
MetS | up-regulated | cholesterol and fatty acid biosynthesis | mice | 115 |
| miRNA-486-5p miRNA-497 miRNA-509-5p miRNA-605 |
MetS | up-regulated | ? | human | 116 |
| miRNA-370-3p miRNA-375 miRNA-15a-5p miRNA-17-5p |
MetS | down-regulated | fatty acid metabolism, AMPK and Wnt signaling pathway | human | 117 |
| miRNA-221 let-7g |
MetS | up-regulated | human | 118 | |
| miRNA-526b-5p miRNA-6516-5p |
MetS | down-regulation | MAPK signaling pathway | human | 119 |
| miRNA-143-3p | MetS | up-regulated | insulin signaling pathway | human | 122 |
| circRNA-H19 | MetS | up-regulated | lipid metabolism | human | 120 |
| lncRNA-XIST | MetS | down-regulated | FoxO, PI3K-AKT & mTOR signaling pathway | human | 121 |
MetS, metabolic syndrome; T2DM, type 2 diabetes mellitus; MSCs, mesenchymal stem cells.
miRNAs in metabolic syndrome
Several studies have reported the role of miRNAs in the pathogenesis of MetS and its involvement in the diagnosis, prognosis, or as a therapeutic target of the disease. miRNA-122 is abundant in the liver and plays a fundamental role in lipid hemostasis. A strong association of circulating miRNA-122 with the risk of developing MetS was reported; circulating miRNA-122 level was significantly increased in MetS and T2DM patients and positively correlated with dyslipidemia. Interestingly, the elevated levels of miRNA-122 occurred before the manifestation of the MetS, rendering it an ideal biomarker for the onset of MetS.111 In a comparative study between lean-mesenchymal stem cells (lean-MSCs) and metabolic syndrome-mesenchymal stem cells (MetS-MSCs), miRNA-196, miRNA-27b, and miRNA-301 were up-regulated in MetS-MSCs compared with Lean-MSCs.112 Goguet-Rubio and his co-researchers reported a significant decrease of miRNA-320a, miRNA-97-3p, miRNA-23-3p, miRNA-221-3p, miRNA-27a-3p, and miRNA-130a-3p in both obesity and MetS patients of the West Virginia population, suggesting a miRNA panel that could be used for the detection of the disease and prevent its progression to the irreversible stage.113 Changes in several miRNA clusters have been reported in MetS patients. Specifically, miRNA-150, miRNA-192, miRNA-27a, miRNA-320a, and miRNA-375 were up-regulated in both MetS and T2DM subjects.114 Additionally, Nasias et al115 reported an increased expression of circulating miRNA-27a and miRNA-130b in high-fat diet-induced MetS mice. In other findings, serum levels of miRNA-486-5p, miRNA-497, miRNA-509-5p and miRNA-605 were significantly up-regulated in MetS patients compared to healthy individuals. Thus, it was suggested that these miRNAs could be considered as early biomarkers and follow-up the prognosis of MetS.116 In another experiment, dysregulation of 26 miRNAs was observed using the RT-qPCR method in individuals with MetS, where miRNA-15a-5p, miRNA-17-5p, miRNA-370-3p, and miRNA-375 were identified as predictive biomarkers of MetS. Further analysis revealed that the expression levels of miRNA-15a-5p and miRNA-17-5p were negatively correlated with various components of MetS including plasma glucose, BMI, waist circumference and blood pressure. Moreover, it showed that the miRNA target genes involved in the regulation of insulin signaling pathway, wnt signaling pathway, TGF-β signaling pathway, AMPK signaling pathway, and fatty acid metabolism, suggesting the role of these miRNAs in the transcriptional regulation associated with the development and progression of complex cardio-metabolic diseases.117 It was discovered that the circulating level of miRNA-221 and let-7g increased significantly in MetS patients as compared to healthy controls. Furthermore, the sex-specific analysis showed that dysregulation was more intense in the female subjects as compared to the males. The elevation of serum let-7g was strongly associated with a low level of HDL-C and high blood pressure.118 Recently, Liu et al119 have reported the down-regulation of miRNA-526b-5p and miRNA-6516-5p in the plasma of MetS patients. Further analysis predicts the involvement of the miRNAs in MetS via the targeting genes of mitogen-activated protein kinase (MAPK), suggesting their potential as biomarkers of the disease.
circRNAs in metabolic syndrome
Despite the number of reports highlighting the crucial role of circRNAs in the individual components of MetS, the literature explaining the direct relationship between circRNAs and MetS is scarce. Recently, Zhu et al120 examined the expression level of circRNA-H19 in the serum of MetS patients. It was found that circRNA-H19 was significantly up-regulated in the MetS subjects and correlated significantly with body mass index, waist circumference, fat percent, visceral fat area (VFA) and HDL-c. The strongest association was observed with the visceral fat area (VFA). circRNA-H19 knock-down promoted hADCSs adipogenic differentiation and lipid accumulation by regulating the expression of PTBP1 and SREBP1. Moreover, multivariate logistic regression analysis revealed a strong association between hsa_circH19 and MetS, independent of gender, age, smoking, drinking, BMI and VFA. The author further ascertained the accuracy and clinical effectiveness of the circ-RNA-H19 using ROC curve analysis and found that the molecule could be used as a diagnostic marker of MetS.
lncRNAs in metabolic syndrome
The involvement of lncRNAs in various components of MetS has been established. However, little is known about the regulatory role of these molecules in MetS. In addition, the direct link between lncRNAs and MetS is not fully elucidated. Recently, MetS-associated lncRNA–miRNA–mRNA network (LMMN) was constructed for the prediction of MetS-associated lncRNAs according to the topological properties of the constructed network and identified lncRNA-XIST as the most important lncRNA in the LMMN network. Further analysis validated the significant decrease in the expression of lncRNA-XIST in MetS patients compared to healthy individuals. Additionally, a negative correlation was observed between the lncRNA-XIST and C-peptide while a positive association was highlighted between the XIST and BMI of MetS patients.121
Thepapeutic potentials of ncRNAs in metabolic syndrome
Microarray data showed that 27 miRNAs were differentially expressed in both serum and urine of patients with MetS compared to healthy individuals. Furthermore, the importance of miRNA-143-3p in the insulin signaling pathway of both MetS patients and HFD induced obese mice was highlighted. The knockdown of miRNA-143-3p in experimental mice prevented the obesity-related IR, via the regulation of insulin-like growth factor 2 receptor (IGF2R). On the other hand, the restoration of the previously knockdown miRNA-143-3p resulted in obesity-related metabolic derangements. It is therefore suggested that inhibition of circulating miRNA-143-3p expression can protect against insulin resistance, which is a fundamental event for the onset of MetS.122 MetS is characterized by an elevated serum level of insulin due to the existence of insulin resistance. This condition results in increased levels of miRNA-33b and its host gene, SREBP1C in the liver, thereby contributing to both elevated VLDL-C levels and a diminished serum level of HDL-C in MetS patient. In its support, a previous study showed that inhibition of miRNA-33a and miRNA-33b leads to a decrease in the plasma level of VLDL-C and increases HDL-C. These results suggest that the development of antagonists of miRNA-33 can be a promising strategy for the therapy of dyslipidaemia associated with MetS and cardiometabolic disorders.123,124 Significant dysregulation of many microRNAs, including miRNA-197, miRNA-23a, miRNA-509-5p, miRNA-130a, miRNA-195, miRNA-27a, and miRNA-320a, has been found to be associated with MetS and the risks involved in the manifestation of the disease. Notably, a credible relationship between miRNA-27a and miRNA-320a with MetS has also been observed. It was therefore suggested that the development of pharmacological agents that suppress the up-regulated miRNAs, e.g., miRNA-197, miRNA-23a, miRNA-509-5p, miRNA-130a, miRNA-195, miRNA-27a, and miRNA-320a, in MetS patients can potentially reinstate the metabolic homeostasis in MetS.114
Based on the literature analysis, miRNA-27, miRNA-221, miRNA-33, and miRNA-320a have been reported frequently to be associated with MetS.112,115,118 As a result, they are worth investigating further to see if they can be used as a diagnostic marker or therapeutic candidate for MetS.
Therapeutic prospects of ncRNAs in metabolic diseases
One of the important properties of ncRNA is its ability to serve as therapeutic agents or therapeutic targets. Unlike conventional man-made chemical based therapy, ncRNAs are endogenous molecules with an established mechanism for their processing in human cells that enables them to execute the therapeutic benefits against a particular disease. In addition, ncRNA acts on several target genes in the target pathway. So, a broader and specific response that may normalize the physiological changes linked to the disease, will consequently be achieved.125 These have conferred them additional advantages over chemical based therapies. Furthermore, a treatment approach that targets ncRNA can efficiently address the molecular basis of the disease. Despite the number of reported preclinical studies on miRNAs, circRNAs, and lncRNAs, only a few have entered clinical trials. Miravirsen is the first anti-miRNA agent that entered to clinical trials. It was designed for the treatment of hepatitis C virus (HCV) infection by targeting miR-122. In its phase II trial (NCT01200420), the effectiveness and safety of miravirsen against chronic HCV genotype 1 infection was commendable.126,127 MRX34 is a liposomal miR-34 mimic that is being tested in a phase I clinical study (NCT01829971) in patients with advanced solid hepatocellular carcinoma tumors.128 However, the trial was halted owing to the immune related side effects.129 Currently, a number of preclinical studies have identified the therapeutic potential of miRNAs, circRNAs, and lncRNAs in various metabolic diseases. In the near future, ncRNA-based intervention will be incorporated into the therapeutic regiment of different metabolic diseases.
Potential challenges of ncRNA-based therapeutics
Despite the availability of small molecules that could effectively inhibit ncRNAs, particularly, miRNA circRNAs and lncRNA, there are still significant obstacles to their clinical application. These include selectivity, optimum potency, delivery, and safety. Off-target effects and inefficient delivery to the location of interest are two hurdles for the development of miRNA-based therapies. Delivery constraints, e.g., degradation by nucleases or inadequate absorption owing to size and negative charge, as well as the hydrophilic nature, cause insufficient delivery of the synthesised oligonucleotides to a specific target to achieve optimal target inhibition.125,129,130 The potential toxicity of the ncRNA-based therapy is also a matter of concern. For instance, administration of miRNA mimics has resulted in the innate immune response associated with the release of IL-6 and TNF, inhibition of coagulation factors, hepatotoxicity, and complement system activation. These off-target effects remain the major hurdles for ncRNA-based therapy.129,130
Conclusion
Circulating ncRNAs function in the regulation of gene expression, their expressional levels change in many metabolic diseases and made them suitable biomarkers for the disease diagnosis, prognosis, or therapeutic target. ncRNAs are involved in the onset of obesity and its progression to MetS and CVD. miRNA-192 miRNA-122 and miRNA-221 were dysregulated in all of these metabolic diseases. Similarly, miRNA-15a, miRNA-26, miRNA-27a, miRNA-320 and miRNA-375 were dysregulated in at least three of the four diseases reviewed. In several cases, knockdown or overexpression of ncRNAs appeared to revert many cellular changes that occur in pathological conditions, this qualified them as a promising molecular target for the treatment or management of the disease. Despite the role of circRNAs in the pathogenesis of many metabolic diseases, there is little or no validated literature elucidating its role in the pathogenesis of MetS. This review will help scientists to explore the diagnostic, prognostic and therapeutic role of ncRNAs in metabolic diseases.
Author contributions
AD performed the literature search and organized the information and drafted the manuscript. MJK initiated and conceived the review and critically revised the manuscript. AN and AL participated in writing and critical review. All authors reviewed and critically revised the manuscript text and approved its final version.
Conflict of interests
Authors declare no conflict of interests.
Consent for publication
All authors are agreed and have consent for publication.
Acknowledgements
The authors acknowledged the Usmanu Danfodiyo University Sokoto, Nigeria for the financial support to AD and COMSATS University Islamabad for providing an enabling environment during the study. Authors are also grateful to Dr. Hassaan Mehboob Awan for his insights and suggestions in data interpretation.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.022.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bayoumi A.S., Aonuma T., Teoh J.P., Tang Y.L., Kim I.M. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol Sin. 2018;39(7):1100–1109. doi: 10.1038/aps.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou W., Bonkovsky H.L. Non-coding RNAs in hepatitis C-induced hepatocellular carcinoma: dysregulation and implications for early detection, diagnosis and therapy. World J Gastroenterol. 2013;19(44):7836–7845. doi: 10.3748/wjg.v19.i44.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling H., Girnita L., Buda O., Calin G.A. Non-coding RNAs: the cancer genome dark matter that matters. Clin Chem Lab Med. 2017;55(5):705–714. doi: 10.1515/cclm-2016-0740. [DOI] [PubMed] [Google Scholar]
- 4.Braicu C., Zimta A.A., Harangus A., et al. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers. 2019;11(5):605. doi: 10.3390/cancers11050605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iaconetti C., Gareri C., Polimeni A., Indolfi C. Non-coding RNAs: the “dark matter” of cardiovascular pathophysiology. Int J Mol Sci. 2013;14(10):19987–20018. doi: 10.3390/ijms141019987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright M.W., Bruford E.A. Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genom. 2011;5(2):90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losko M., Kotlinowski J., Jura J. Long noncoding RNAs in metabolic syndrome related disorders. Mediat Inflamm. 2016;2016 doi: 10.1155/2016/5365209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su Y., Wu H., Pavlosky A., et al. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis. 2016;7(8) doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arraiano C.M. Regulatory noncoding RNAs: functions and applications in health and disease. FEBS J. 2021;288(22):6308–6309. doi: 10.1111/febs.16027. [DOI] [PubMed] [Google Scholar]
- 10.Lekka E., Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soni D.K., Biswas R. Role of non-coding RNAs in post-transcriptional regulation of lung diseases. Front Genet. 2021;12 doi: 10.3389/fgene.2021.767348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu M., Chen J., Tao Z., et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76(3):441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859(1):163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 15.He X., Ou C., Xiao Y., Han Q., Li H., Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8(41):71325–71341. doi: 10.18632/oncotarget.19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Zhao Z., Wang M., Zhou X. London IntechOpen; 2019. The Role of Long Noncoding RNAs in Gene Expression Regulation; pp. 1–17. (Gene Expression Profiling in Cancer). [Google Scholar]
- 17.Choquet H., Meyre D. Molecular basis of obesity: current status and future prospects. Curr Genom. 2011;12(3):154–168. doi: 10.2174/138920211795677921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabir J.S.M., El Omri A., Banaganapalli B., et al. Unraveling the role of salt-sensitivity genes in obesity with integrated network biology and co-expression analysis. PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0228400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pescador N., Pérez-Barba M., Ibarra J.M., Corbatón A., Martínez-Larrad M.T., Serrano-Ríos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo W., Esguerra J.L.S., Eliasson L., Melander O. miR-483-5p associates with obesity and insulin resistance and independently associates with new onset diabetes mellitus and cardiovascular disease. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0206974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghorbani S., Mahdavi R., Alipoor B., et al. Decreased serum microRNA-21 level is associated with obesity in healthy and type 2 diabetic subjects. Arch Physiol Biochem. 2018;124(4):300–305. doi: 10.1080/13813455.2017.1396349. [DOI] [PubMed] [Google Scholar]
- 22.Al-Rawaf H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin Nutr. 2019;38(5):2231–2238. doi: 10.1016/j.clnu.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Cui X., You L., Zhu L., et al. Change in circulating microRNA profile of obese children indicates future risk of adult diabetes. Metabolism. 2018;78:95–105. doi: 10.1016/j.metabol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Castaño C., Kalko S., Novials A., Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115(48):12158–12163. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGregor R.A., Choi M.S. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F., Ma D., Zhao W., et al. Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat Commun. 2020;11(1):1822. doi: 10.1038/s41467-020-15529-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Church R.J., Kullak-Ublick G.A., Aubrecht J., et al. Candidate biomarkers for the diagnosis and prognosis of drug-induced liver injury: an international collaborative effort. Hepatology. 2019;69(2):760–773. doi: 10.1002/hep.29802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subudhi S., Drescher H.K., Dichtel L.E., et al. Distinct hepatic gene-expression patterns of NAFLD in patients with obesity. Hepatol Commun. 2022;6(1):77–89. doi: 10.1002/hep4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonsembiante L., Targher G., Maffeis C. Non-alcoholic fatty liver disease in obese children and adolescents: a role for nutrition? Eur J Clin Nutr. 2022;76(1):28–39. doi: 10.1038/s41430-021-00928-z. [DOI] [PubMed] [Google Scholar]
- 30.Fang Z., Dou G., Wang L. MicroRNAs in the pathogenesis of nonalcoholic fatty liver disease. Int J Biol Sci. 2021;17(7):1851–1863. doi: 10.7150/ijbs.59588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H., Tian Y., Tang D., et al. An endoplasmic reticulum stress-microRNA-26a feedback circuit in NAFLD. Hepatology. 2021;73(4):1327–1345. doi: 10.1002/hep.31428. [DOI] [PubMed] [Google Scholar]
- 32.Zaiou M. The emerging role and promise of circular RNAs in obesity and related metabolic disorders. Cells. 2020;9(6):1473. doi: 10.3390/cells9061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Sulaiti H., Dömling A.S., Elrayess M.A. IntechOpen; 2019. Mediators of Impaired Adipogenesis in Obesity-Associated Insulin Resistance and T2DM. Adipose Tissue - an Update; pp. 1–26. [Google Scholar]
- 34.Li A., Huang W., Zhang X., Xie L., Miao X. Identification and characterization of CircRNAs of two pig breeds as a new biomarker in metabolism-related diseases. Cell Physiol Biochem. 2018;47(6):2458–2470. doi: 10.1159/000491619. [DOI] [PubMed] [Google Scholar]
- 35.Arcinas C., Tan W., Fang W., et al. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab. 2019;1(7):688–703. doi: 10.1038/s42255-019-0078-z. [DOI] [PubMed] [Google Scholar]
- 36.Sun W., Sun X., Chu W., Yu S., Dong F., Xu G. CircRNA expression profiles in human visceral preadipocytes and adipocytes. Mol Med Rep. 2020;21(2):815–821. doi: 10.3892/mmr.2019.10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y., Liu H., Li Y., et al. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics. 2020;10(10):4705–4719. doi: 10.7150/thno.42417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J., Ruan Y., Wang M., et al. Differentially expressed circulating LncRNAs and mRNA identified by microarray analysis in obese patients. Sci Rep. 2016;6 doi: 10.1038/srep35421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu B., Gerin I., Miao H., et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Xue C., Lin J., et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci Transl Med. 2018;10(446) doi: 10.1126/scitranslmed.aar5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowak A., Kobierzycki C., Dziegiel P. The Role of leptin in pathogenesis of obesity-related cancers. Postepy Biol Komorki. 2015;42(2):309–328. [Google Scholar]
- 42.Izquierdo A.G., Crujeiras A.B., Casanueva F.F., Carreira M.C. Leptin, obesity, and leptin resistance: where are we 25 years later? Nutrients. 2019;11(11):2704. doi: 10.3390/nu11112704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo K.A., Huang S., Walet A.C.E., et al. Adipocyte long-noncoding RNA transcriptome analysis of obese mice identified lnc-leptin, which regulates leptin. Diabetes. 2018;67(6):1045–1056. doi: 10.2337/db17-0526. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F.F., Liu Y.H., Wang D.W., et al. Obesity-induced reduced expression of the lncRNA ROIT impairs insulin transcription by downregulation of Nkx 6.1 methylation. Diabetologia. 2020;63(4):811–824. doi: 10.1007/s00125-020-05090-y. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y., Ji Y., Li M., et al. Integrated analysis of long noncoding RNA and mRNA expression profile in children with obesity by microarray analysis. Sci Rep. 2018;8(1):8750. doi: 10.1038/s41598-018-27113-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esau C., Davis S., Murray S.F., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabol. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., He Y., Mackowiak B., Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70(4):784–795. doi: 10.1136/gutjnl-2020-322526. [DOI] [PubMed] [Google Scholar]
- 48.He Y., Rodrigues R.M., Wang X., et al. Neutrophil-to-hepatocyte communication via LDLR-dependent miR-223-enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. J Clin Invest. 2021;131(3):141513–151415. doi: 10.1172/JCI141513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Natarajan R., Putta S., Kato M. MicroRNAs and diabetic complications. J Cardiovasc Transl Res. 2012;5(4):413–141549. doi: 10.1007/s12265-012-9368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiménez-Lucena R., Camargo A., Alcalá-Diaz J.F., et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: from the CORDIOPREV study. Exp Mol Med. 2018;50(12):1–12. doi: 10.1038/s12276-018-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiménez-Lucena R., Rangel-Zúñiga O.A., Alcalá-Díaz J.F., et al. Circulating miRNAs as predictive biomarkers of type 2 diabetes mellitus development in coronary heart disease patients from the CORDIOPREV study. Mol Ther Nucleic Acids. 2018;12:146–157. doi: 10.1016/j.omtn.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Muhtaresh H.A., Al-Kafaji G. Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. J Clin Med. 2018;7(2):12. doi: 10.3390/jcm7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng X., Liu Y., Luo M., et al. Circulating miRNA-24 and its target YKL-40 as potential biomarkers in patients with coronary heart disease and type 2 diabetes mellitus. Oncotarget. 2017;8(38):63038–63046. doi: 10.18632/oncotarget.18593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaeger A., Zollinger L., Saely C.H., et al. Circulating microRNAs-192 and-194 are associated with the presence and incidence of diabetes mellitus. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-32274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witkowski M., Weithauser A., Tabaraie T., et al. Micro-RNA-126 reduces the blood thrombogenicity in diabetes mellitus via targeting of tissue factor. Arterioscler Thromb Vasc Biol. 2016;36(6):1263–1271. doi: 10.1161/ATVBAHA.115.306094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannella A., Radu C.M., Franco L., et al. Circulating levels and characterization of microparticles in patients with different degrees of glucose tolerance. Cardiovasc Diabetol. 2017;16(1):118. doi: 10.1186/s12933-017-0600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stępień E.Ł., Durak-Kozica M., Kamińska A., et al. Circulating ectosomes: determination of angiogenic microRNAs in type 2 diabetes. Theranostics. 2018;8(14):3874–3890. doi: 10.7150/thno.23334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dooley J., Garcia-Perez J.E., Sreenivasan J., et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes. 2016;65(1):53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satake E., Pezzolesi M.G., Md Dom Z.I., Smiles A.M., Niewczas M.A., Krolewski A.S. Circulating miRNA profiles associated with hyperglycemia in patients with type 1 diabetes. Diabetes. 2018;67(5):1013–1023. doi: 10.2337/db17-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornfeld J.W., Baitzel C., Könner A.C., et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 61.Zhao C., Dong J., Jiang T., et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collares C.V., Evangelista A.F., Xavier D.J., et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:491. doi: 10.1186/1756-0500-6-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Regmi A., Liu G., Zhong X., et al. Evaluation of serum microRNAs in patients with diabetic kidney disease: a nested case-controlled study and bioinformatics analysis. Med Sci Mon Int Med J Exp Clin Res. 2019;25:1699–1708. doi: 10.12659/MSM.913265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan L., Lian W., Zhang X., et al. Human circular RNA-0054633 regulates high glucose-induced vascular endothelial cell dysfunction through the microRNA-218/roundabout 1 and microRNA-218/heme oxygenase-1 axes. Int J Mol Med. 2018;42(1):597–606. doi: 10.3892/ijmm.2018.3625. [DOI] [PubMed] [Google Scholar]
- 65.Wu H., Wu S., Zhu Y., et al. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin Epigenet. 2019;11(1):22. doi: 10.1186/s13148-019-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Z., Li X., Jian D., Hao P., Rao L., Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haddad G., Lorenzen J.M. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428. doi: 10.3389/fphar.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haque S., Ames R.M., Moore K., Lee B.P., Jeffery N., Harries L.W. Islet-expressed circular RNAs are associated with type 2 diabetes status in human primary islets and in peripheral blood. BMC Med Genom. 2020;13(1):64. doi: 10.1186/s12920-020-0713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fang Y., Wang X., Li W., et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 2018;42(4):1865–1874. doi: 10.3892/ijmm.2018.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu W., Han Q., Zhao L., Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol. 2019;234(2):1469–1476. doi: 10.1002/jcp.26959. [DOI] [PubMed] [Google Scholar]
- 71.Carter G., Miladinovic B., Patel A.A., Deland L., Mastorides S., Patel N.A. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015;4:102–107. doi: 10.1016/j.bbacli.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge X., Xu B., Xu W., et al. Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging (Albany NY) 2019;11(20):8745–8759. doi: 10.18632/aging.102249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X., Zeng L., Cao C., et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res. 2017;350(2):327–335. doi: 10.1016/j.yexcr.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 74.Abdulle L.E., Hao J.L., Pant O.P., et al. MALAT1 as a diagnostic and therapeutic target in diabetes-related complications: a promising long-noncoding RNA. Int J Med Sci. 2019;16(4):548–555. doi: 10.7150/ijms.30097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jo S., Chen J., Xu G., Grayson T.B., Thielen L.A., Shalev A. miR-204 controls glucagon-like peptide 1 receptor expression and agonist function. Diabetes. 2018;67(2):256–264. doi: 10.2337/db17-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fu X., Jin L., Wang X., et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci U S A. 2013;110(44):17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu X., Dong B., Tian Y., et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125(6):2497–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu H., Du X., Xu J., et al. Pancreatic β cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving β cell function. PLoS Biol. 2020;18(2) doi: 10.1371/journal.pbio.3000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu Y., Ke G., Wang L., Zhou E., Zhu K., Wei Y. Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res. 2017;58(3):176–184. doi: 10.1159/000479156. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Z., Gao W., Long Q.Q., et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7(1):7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marchetti P., Bugliani M., De Tata V., Suleiman M., Marselli L. Pancreatic beta cell identity in humans and the role of type 2 diabetes. Front Cell Dev Biol. 2017;5:55. doi: 10.3389/fcell.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alimoradi N., Firouzabadi N., Fatehi R. Metformin and insulin-resistant related diseases: emphasis on the role of microRNAs. Biomed Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111662. [DOI] [PubMed] [Google Scholar]
- 83.Skuratovskaia D., Vulf M., Komar A., Kirienkova E., Litvinova L. Promising directions in atherosclerosis treatment based on epigenetic regulation using microRNAs and long noncoding RNAs. Biomolecules. 2019;9(6):226. doi: 10.3390/biom9060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang Y., Sun-Waterhouse D., Chen Y., Li F., Li D. Epigenetic mechanisms underlying the benefits of flavonoids in cardiovascular health and diseases: are long non-coding RNAs rising stars? Crit Rev Food Sci Nutr. 2022;62(14):3855–3872. doi: 10.1080/10408398.2020.1870926. [DOI] [PubMed] [Google Scholar]
- 85.Michell D.L., Vickers K.C. HDL and microRNA therapeutics in cardiovascular disease. Pharmacol Ther. 2016;168:43–52. doi: 10.1016/j.pharmthera.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gangwar R.S., Rajagopalan S., Natarajan R., Deiuliis J.A. Noncoding RNAs in cardiovascular disease: pathological relevance and emerging role as biomarkers and therapeutics. Am J Hypertens. 2018;31(2):150–165. doi: 10.1093/ajh/hpx197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang Q.J., Lei H.P., Wu H., et al. Plasma miR-142 predicts major adverse cardiovascular events as an intermediate biomarker of dual antiplatelet therapy. Acta Pharmacol Sin. 2019;40(2):208–215. doi: 10.1038/s41401-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Widera C., Gupta S.K., Lorenzen J.M., et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51(5):872–875. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 89.Cavarretta E., Frati G. MicroRNAs in coronary heart disease: ready to enter the clinical arena? BioMed Res Int. 2016;2016 doi: 10.1155/2016/2150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Navickas R., Gal D., Laucevičius A., Taparauskaitė A., Zdanytė M., Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karakas M., Schulte C., Appelbaum S., et al. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur Heart J. 2017;38(7):516–523. doi: 10.1093/eurheartj/ehw250. [DOI] [PubMed] [Google Scholar]
- 92.Kaudewitz D., Zampetaki A., Mayr M. MicroRNA biomarkers for coronary artery disease? Curr Atherosclerosis Rep. 2015;17(12):70. doi: 10.1007/s11883-015-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y.L., Yu W. Association of circulating microRNA-122 with presence and severity of atherosclerotic lesions. PeerJ. 2018;6 doi: 10.7717/peerj.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jansen F., Yang X., Proebsting S., et al. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc. 2014;3(6) doi: 10.1161/JAHA.114.001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J., Pei Y., Zhong Y., Jiang S., Shao J., Gong J. Altered serum microRNAs as novel diagnostic biomarkers for atypical coronary artery disease. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jakobi T., Siede D., Eschenbach J., et al. Deep characterization of circular RNAs from human cardiovascular cell models and cardiac tissue. Cells. 2020;9(7):1616. doi: 10.3390/cells9071616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holdt L.M., Stahringer A., Sass K., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7 doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geng H.H., Li R., Su Y.M., et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Z., Li X., Gao C., et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7 doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu T., Wu D., Wu Q., et al. Knockdown of long non-coding RNA-ZFAS1 protects cardiomyocytes against acute myocardial infarction via anti-apoptosis by regulating miR-150/CRP. J Cell Biochem. 2017;118(10):3281–3289. doi: 10.1002/jcb.25979. [DOI] [PubMed] [Google Scholar]
- 101.Josefs T., Boon R.A. The long non-coding road to atherosclerosis. Curr Atherosclerosis Rep. 2020;22(10):55. doi: 10.1007/s11883-020-00872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H., Han S., Sun Q., et al. Long non-coding RNA CDKN2B-AS1 reduces inflammatory response and promotes cholesterol efflux in atherosclerosis by inhibiting ADAM10 expression. Aging (Albany NY) 2019;11(6):1695–1715. doi: 10.18632/aging.101863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Z., Ye P., Wang S., et al. MicroRNA-150 protects the heart from injury by inhibiting monocyte accumulation in a mouse model of acute myocardial infarction. Circ Cardiovasc Genet. 2015;8(1):11–20. doi: 10.1161/CIRCGENETICS.114.000598. [DOI] [PubMed] [Google Scholar]
- 104.Theodorou K., Boon R.A. Endothelial cell metabolism in atherosclerosis. Front Cell Dev Biol. 2018;6:82. doi: 10.3389/fcell.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feinberg M.W., Moore K.J. MicroRNA regulation of atherosclerosis. Circ Res. 2016;118(4):703–720. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meiler S., Baumer Y., Toulmin E., Seng K., Boisvert W.A. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35(2):323–331. doi: 10.1161/ATVBAHA.114.304878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meng X.D., Yao H.H., Wang L.M., et al. Knockdown of GAS5 inhibits atherosclerosis progression via reducing EZH2-mediated ABCA1 transcription in ApoE-/- mice. Mol Ther Nucleic Acids. 2020;19:84–96. doi: 10.1016/j.omtn.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pan J.X. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(2):322–328. [PubMed] [Google Scholar]
- 109.Zhuang Y., Li T., Xiao H., et al. LncRNA-H19 drives cardiomyocyte senescence by targeting miR-19a/socs 1/p53 axis. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.631835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ranasinghe P., Mathangasinghe Y., Jayawardena R., Hills A.P., Misra A. Prevalence and trends of metabolic syndrome among adults in the Asia-Pacific region: a systematic review. BMC Publ Health. 2017;17(1):101. doi: 10.1186/s12889-017-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Willeit P., Skroblin P., Moschen A.R., et al. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes. 2017;66(2):347–357. doi: 10.2337/db16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meng Y., Eirin A., Zhu X.Y., et al. Micro-RNAS regulate metabolic syndrome-induced senescence in porcine adipose tissue-derived mesenchymal stem cells through the P16/MAPK pathway. Cell Transplant. 2018;27(10):1495–1503. doi: 10.1177/0963689718795692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goguet-Rubio P., Klug R.L., Sharma D.L., et al. Existence of a strong correlation of biomarkers and miRNA in females with metabolic syndrome and obesity in a population of West Virginia. Int J Med Sci. 2017;14(6):543–553. doi: 10.7150/ijms.18988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karolina D.S., Tavintharan S., Armugam A., et al. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97(12):E2271–E2276. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 115.Nasias D., Evangelakos I., Nidris V., et al. Significant changes in hepatic transcriptome and circulating miRNAs are associated with diet-induced metabolic syndrome in apoE3L.CETP mice. J Cell Physiol. 2019;234(11):20485–20500. doi: 10.1002/jcp.28649. [DOI] [PubMed] [Google Scholar]
- 116.Bakr Zaki M., Abulsoud A.I., Elsisi A.M., et al. Potential role of circulating microRNAs (486-5p, 497, 509-5p and 605) in metabolic syndrome Egyptian male patients. Diabetes Metab Syndr Obes. 2019;12:601–611. doi: 10.2147/DMSO.S187422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramzan F., D'Souza R.F., Durainayagam B.R., et al. Circulatory miRNA biomarkers of metabolic syndrome. Acta Diabetol. 2020;57(2):203–214. doi: 10.1007/s00592-019-01406-6. [DOI] [PubMed] [Google Scholar]
- 118.Wang Y.T., Tsai P.C., Liao Y.C., Hsu C.Y., Juo S.H.H. Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci. 2013;20(1):72. doi: 10.1186/1423-0127-20-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu G., Lei Y., Luo S., et al. MicroRNA expression profile and identification of novel microRNA biomarkers for metabolic syndrome. Bioengineered. 2021;12(1):3864–3872. doi: 10.1080/21655979.2021.1952817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu Y., Gui W., Lin X., Li H. Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp Cell Res. 2020;387(2) doi: 10.1016/j.yexcr.2019.111753. [DOI] [PubMed] [Google Scholar]
- 121.Yao D., Lin Z., Zhan X., Zhan X. Identifying potential functional lncRNAs in metabolic syndrome by constructing a lncRNA-miRNA-mRNA network. J Hum Genet. 2020;65(11):927–938. doi: 10.1038/s10038-020-0753-7. [DOI] [PubMed] [Google Scholar]
- 122.Xihua L., Shengjie T., Weiwei G., et al. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl Res. 2019;205:33–43. doi: 10.1016/j.trsl.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 123.Rayner K.J., Esau C.C., Hussain F.N., et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rottiers V., Näär A.M. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baek J., Kang S., Min H. MicroRNA-targeting therapeutics for hepatitis C. Arch Pharm Res (Seoul) 2014;37(3):299–305. doi: 10.1007/s12272-013-0318-9. [DOI] [PubMed] [Google Scholar]
- 127.Takahashi M., Han S.P., Scherer L.J., Yoon S., Rossi J.J. Elsevier; Amsterdam: 2017. Current Progress and Future Prospects in Nucleic Acid Based Therapeutics. Comprehensive Medicinal Chemistry III; pp. 280–313. [Google Scholar]
- 128.Beg M.S., Brenner A.J., Sachdev J., et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest N Drugs. 2017;35(2):180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roy S., Trautwein C., Luedde T., Roderburg C. A general overview on non-coding RNA-based diagnostic and therapeutic approaches for liver diseases. Front Pharmacol. 2018;9:805. doi: 10.3389/fphar.2018.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baumann V., Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014;6(17):1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.