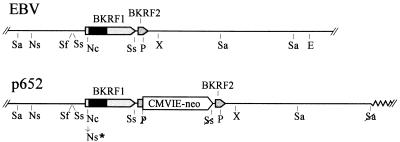
FIG. 1.
Insertion of a selective gene, CMVIE-neo, next to the EBNA-1 gene without disrupting the adjacent gene, BKRF2. (Upper) Part of the EBV genome, from the SalI site at 105,296 to the EcoRV site at 116,865. The EBNA-1 coding sequence (BKRF1) and an adjacent gene, BKRF2, are indicated. The dark portion of the EBNA-1 coding region is IR3, which encodes the Gly-Gly-Ala repeats. The indicated restriction enzyme sites are as follows: E, EcoRV; P, PvuII; Nc, NcoI; Ns, NsiI; Sa, SalI; Sf, SfiI; Ss, SstII; X, XbaI. Only the most relevant sites are shown for NcoI, PvuII, and SstII. (Lower) Relevant portion of the plasmid p652. The CMVIE-neo gene was inserted along with a duplication of nearly 400 bp of EBV sequence, from an SstII site to a PvuII site, placing the duplicated sequence on each side of the insertion. The duplicated sequence includes the 3′ end of the EBNA-1 gene (BKRF1) and continues through much of the adjacent gene, BKRF2. Thus a complete copy of each gene together with its 3′ and 5′ regulatory regions remains intact. Restriction sites that were destroyed where sequences were joined are indicated with a slash. Ns∗ represents filling in of an NcoI site to create an NsiI site and a frameshift mutation in the EBNA-1 gene. The zig-zag line at the right end indicates plasmid vector sequences.