Extended Data Fig. 5 |. Transcriptional profiling of pediatric cardiac fibroblasts and endothelial cells.
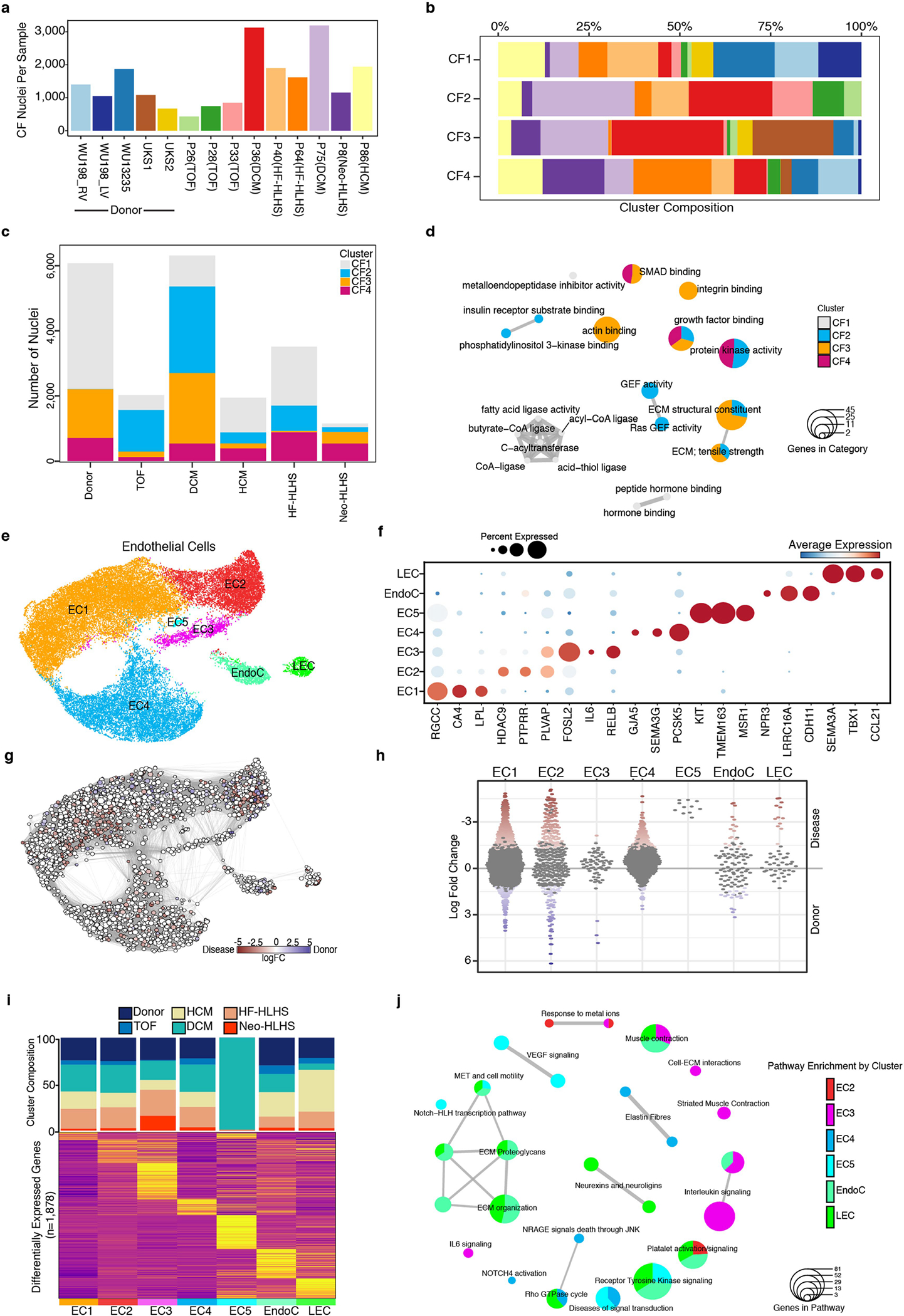
a, Bar plot indicating the number of fibroblast nuclei detected from each sample. b, Stacked bar plot showing the composition of each sample across the indicated CF clusters. c, Stacked bar graph displaying the total number of detected CF nuclei from each patient group. The composition of each CF cluster is highlighted. d, Enrichment map for gene pathway overrepresentation analysis colored by CF cluster. e, UMAP manifold of cardiac ECs colored by cluster. (f) Dot plot of snRNA-seq expression for EC marker genes. g, Embedding of the Milo K-NN differential abundance testing results for ECs. All nodes represent neighborhoods, colored by their log fold changes for disease versus donor. Neighborhoods with insignificant log fold changes (FDR 10%) are white. Layout of nodes determined by UMAP embedding, shown in Extended Data Fig. 5e. h, Beeswarm plot showing the log-fold distribution of changes across disease and donors in neighborhoods from different EC clusters. Differentially abundant neighborhoods are shown in color. i, Top, cluster composition bar plot colored by patient diagnosis. Bottom, heatmap displaying average expression for all differentially expressed genes for EC clusters. j, Enrichment map for gene pathway over-representation analysis colored by EC cluster.
