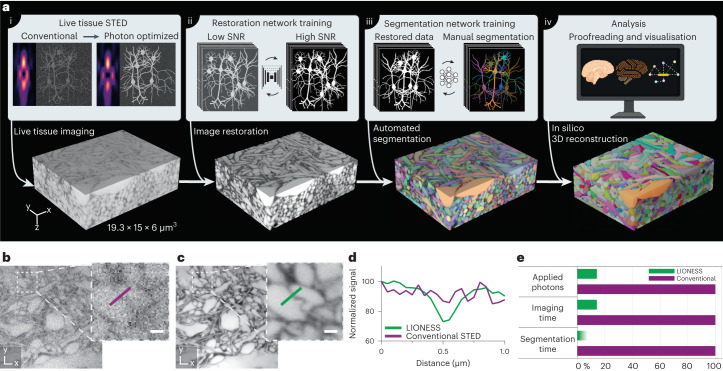
Fig. 1. LIONESS enables dense reconstruction of living brain tissue.
a, LIONESS technology exemplified in living human cerebral organoid. Optical improvements, deep-learning training and analysis (top) flow into individual processing steps (bottom). (i) Near-infrared STED with light patterns for improved effective point-spread-function in tissue. (ii) Deep neural network training on paired low-exposure, low-SNR and high-exposure, high-SNR 3D super-resolved volumes recorded in extracellularly labeled tissue. (iii) Deep 3D-segmentation network training with manually annotated data. (iv) Postprocessing. b, Conventional STED imaging in CA1 neuropil of extracellularly labeled organotypic hippocampal slice culture with phase modulation patterns for lateral (xy) plus axial (z)-resolution increase. c, Same region imaged in LIONESS mode with tissue-adapted STED patterns (4π-helical plus π-top-hat phase modulation), modified detector setup and deep-learning-based image restoration. STED power and dwell time were identical in b and c. The images are representative of n = 3 technical replicates from two samples. Scale bars, 500 nm. d, Line profiles across a putative synaptic cleft as indicated in b and c. e, Schematic comparison of LIONESS imaging with conventional STED imaging (based on the parameters used in restoration network training) in terms of light exposure and imaging time, as well as the reduction in segmentation time by automated over manual segmentation. The shading indicates that reduction in segmentation time by deep learning depends on sample complexity. LIONESS lookup tables are linear and inverted throughout, ranging from black (maximum photon counts extracellularly) to white.