Fig. 1. Conceptual model illustration in which input data (top) consist of chromatin accessibility (ATAC), gene expression (RNA) or both data types (multiome).
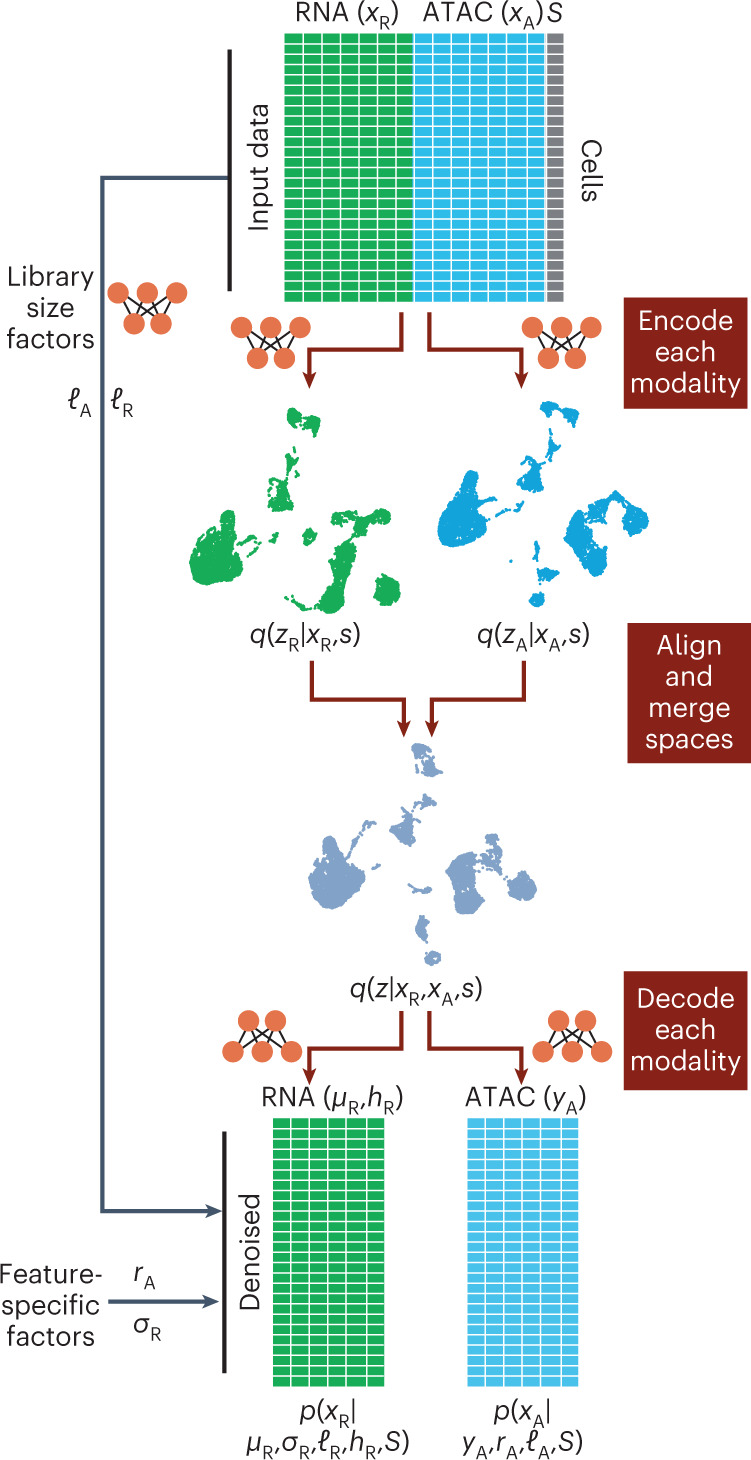
Variable S represents experimental covariates, such as batch or experimental condition. Each data modality is encoded into modality-independent latent representations (using neural network encoders) and then, these representations are merged into a joint latent space. The joint latent representation is used to estimate (decode) the input data together with chromatin region-specific effects (rA), gene-specific dispersion (σR), cell-specific effects (ℓA, ℓR), accessibility probability estimates (YZ) and mean gene expression values (μR).
