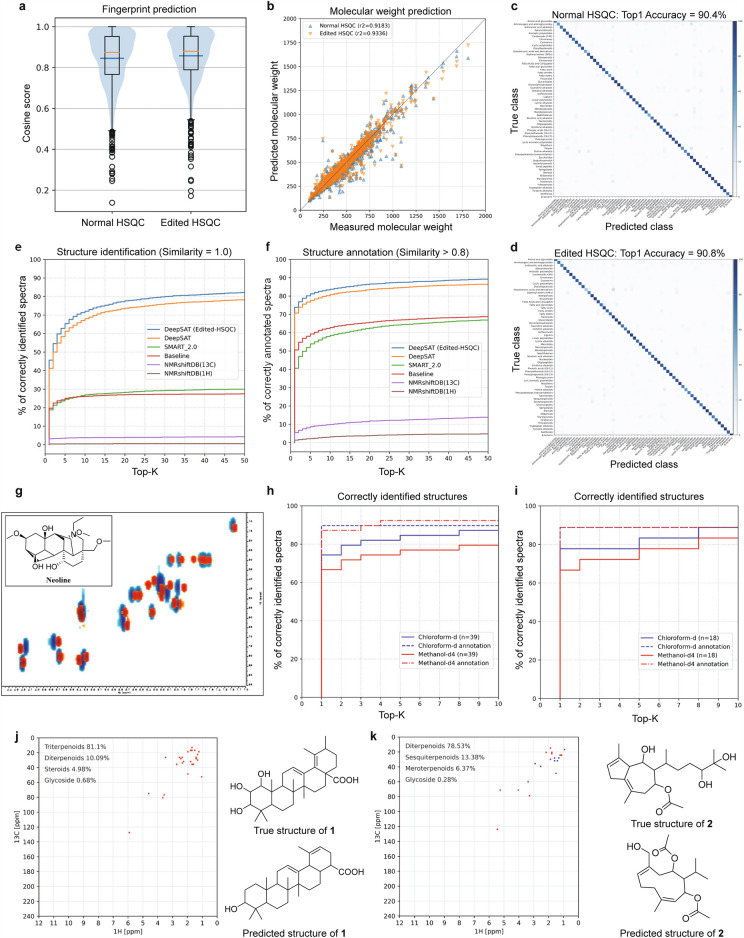
Fig. 2.
Evaluation of DeepSAT using a test set. a Average (orange line) and median (blue line) of cosine scores between predicted and ground truth fingerprints for HSQC and Edited HSQC data input. b Linear regression between measured (x axis) and predicted molecular weights (y axis). c and d Confusion matrix of classification results using DeepSAT with normal HSQC data and multiplicity edited HSQC data. e Percentage of correctly identified structures in the top k output of several different tools, for maximum rank k = 1, 2, …, 50. f Percent of correctly annotated structures in the top k. For the measurement of annotation rate, cosine score of 0.8 was set as the threshold. g Experimental HSQC spectrum of the natural product, neoline dissolved in chloroform-d (blue) and methanol-d4 (red). h Identification (solid) and annotation (dashed) rates in total experimental data. i Identification (solid) and annotation (dashed) rates in compounds with NMR data recorded in both solvents. j and k HSQC spectra and predicted results of previously undescribed compounds 1 and 2