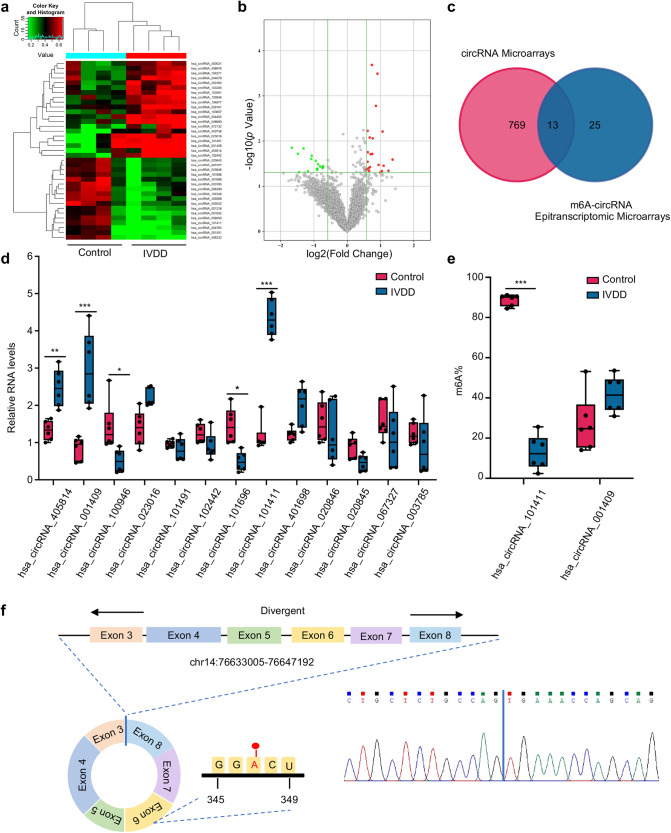
Fig. 1. circGPATCH2L is hypomethylated and upregulated in IVDD.
a Hierarchical clustering for circRNAs with differential m6A methylation levels in the IVDD and control groups. The red–green gradient colour scheme represents the high and low m6A methylation relative levels as referenced in the Colour Key. b Volcano plot showing 38 differentially m6A-methylated circRNAs in NP tissues of the IVDD and control groups. The cut off is |fold change| ≥ 1.5, p ≤ 0.05. The red and green points in the plot indicate the upregulated and downregulated circRNAs with statistical significance, respectively. c Venn diagram identifying 13 differentially expressed circRNAs with altered methylation in IVDD samples based on the overlap of circRNA microarray and m6A-circRNA epitranscriptomic microarray data. d qRT–PCR analysis of the expression of 13 circRNAs in NP tissues. n = 6. e The m6A ratio of hsa_circRNA_101411 and hsa_circRNA_001409 was calculated using a T3 DNA ligase assay. n = 6. f Schematic of the genomic loci of circGPATCH2L and the m6A site. The back-splice junction site of circGPATCH2L was confirmed by Sanger sequencing. NP nucleus pulposus, m6A N6-methyladenosine, qRT–PCR quantitative real-time polymerase chain reaction, IVDD intervertebral disc degeneration. Data are shown as the mean ± S.D. ***p < 0.001; **p < 0.01; *p < 0.05.