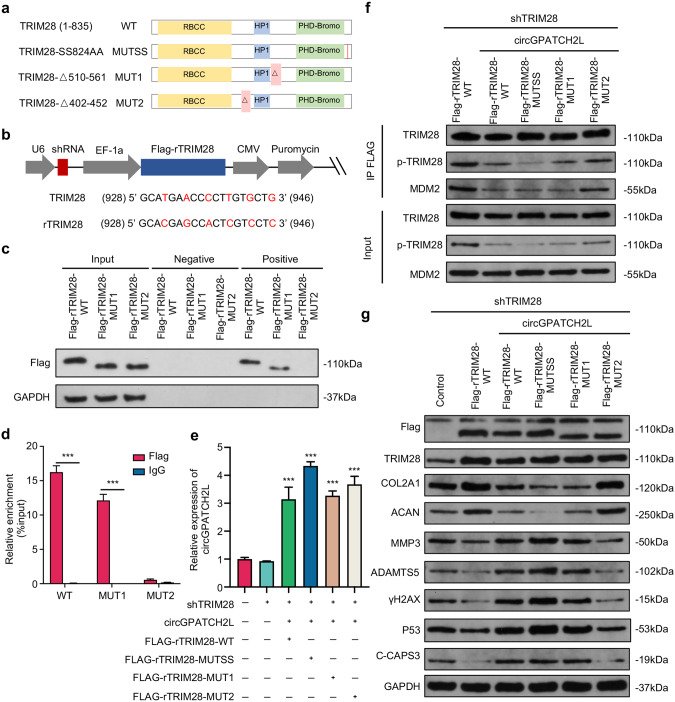
Fig. 4. circGPATCH2L functions in NPCs by binding the aa 402–452 region of TRIM28.
a Schematic representation of the full-length, truncated or point mutant TRIM28. TRIM28 wild type (TRIM28-WT) was full-length and contained three main TRIM28 domains. TRIM28-MUT1 and TRIM28-MUT2 lacked the aa 510–561 and aa 402–452 regions, respectively. Each serine was mutated to alanine at aa 823/824 (TRIM28-MUTSS) to avoid the possibility that Ser823 becomes phosphorylated when Ser824 is mutated. b Schematic diagrams of the lentiviral plasmids used in this study. The modified plasmid was designed for simultaneous expression of shTRIM28, Flag-tagged shTRIM28-resistant wild-type or mutant TRIM28 cDNA (rTRIM28), and the puromycin resistance gene. The shRNA-targeted sequences and the silencing mutations in shTRIM28-resistant cDNAs are shown below. The numbers correspond to the nt positions in the open reading frames of TRIM28, and the changed bases are marked in red. c, d RNA pulldown and RIP assays confirmed the interaction between circGPATCH2L and TRIM28 within the site at residues 402–452. c Coprecipitation of circGPATCH2L and wild-type or mutant TRIM28 in an RNA pulldown assay. Total protein (Input), precipitates with biotinylated probes targeting the circGPATCH2L back-spliced site (positive) or negative biotinylated probes (negative) were analysed via western blot with Flag antibody. d RIP experiments were performed using Flag or negative IgG antibody. Purified RNA was used for qRT–PCR assays. n = 3. e qRT–PCR analysis confirmed the overexpression efficiency of circGPATCH2L in NPCs after treatment with the circGPATCH2L overexpression plasmid in different groups. n = 3. f Cell lysates were immunoprecipitated with an antibody against Flag and analysed via western blot with TRIM28 antibody, p-TRIM28 antibody, or MDM2 antibody (top). Total protein from cell lysates was analysed as input (bottom). g Western blot analysis of Flag, TRIM28, COL2, ACAN, MMP3, ADAMTS5, γH2AX, P53, C-CAPS3, and GAPDH expression in NPCs. The control group was transfected with a blank vector with 3×FLAG. NPCs nucleus pulposus cells, qRT–PCR quantitative real-time polymerase chain reaction, TRIM28 tripartite motif containing 28, p-TRIM28 TRIM28 phosphorylation, MDM2 E3 ubiquitin-protein ligase Mdm2, γH2AX H2A.X variant histone phosphorylation, P53 cellular tumour antigen p53, COL2A1 collagen type II alpha 1 chain, ACAN aggrecan, MMP3 matrix metallopeptidase 3, ADAMTS5 a disintegrin and metalloproteinase with thrombospondin type 1 motif 5, C-CASP3 cleaved Caspase 3, GAPDH glyceraldehyde 3-phosphate dehydrogenase. Data are shown as the mean ± S.D. ***p < 0.001.