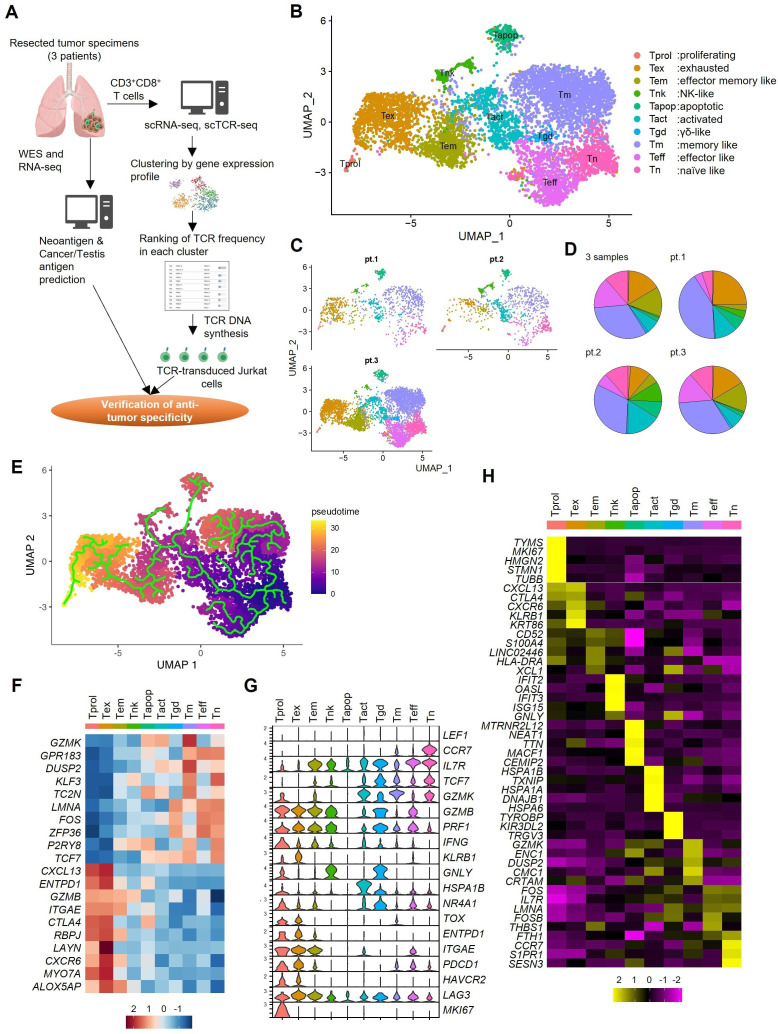
Figure 1.
Phenotypic clustering of CD8+ TILs by single-cell RNA sequencing. (A) Overall scheme of this study. (B) The Uniform Manifold Approximation and Projection (UMAP) of the expression profiles of the 6998 single CD8+ T cells derived from the three surgical lung tumor tissues. CD8+ T cells are classified into 10 distinct transcriptional clusters. (C) UMAPs of CD8+ T cells in each patient. (D) Pie charts showing the proportions of CD8+ T cells in each cluster for all three samples and each patient separately. (E) Pseudotime trajectory analysis of 6998 CD8+ T cells. Each dot represents one single cell and each cell with a pseudotime score from dark blue to yellow, indicating early and terminal states, respectively. (F) The normalized average expression of phenotypic and functional signatures for CD8+ T cells subpopulations defined in B. (G) Violin plots quantifying relative transcriptional expression of genes (rows) with high differential expression among each cluster (columns). (H) Relative expression of the top five most differentially expressed genes in each cluster. NK, natural killer; scRNA-seq, single cell RNA sequencing; scCR-seq, single cell TCR sequencing; Tact, activated T; TCR, T-cell receptor; Tem, effector memory T; Tex, exhausted T; Tm, memory T; Tn, naive T; Tprol, proliferating T; Teff, effector like T; Tgd, γδ-like T; Tapop, apoptotic T; WES, whole exome sequencing.