Figure 2. Real‐time nanopore sequencing technology.
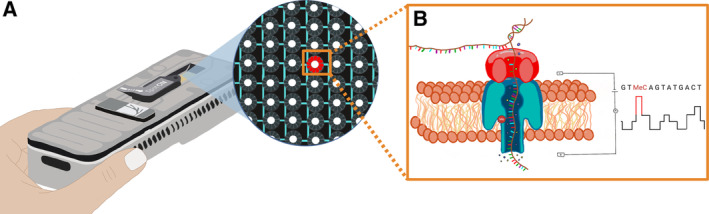
(A) The portable nanopore sequencing device MinION (version Mk1b); the disposable flow cell consists of a fluid‐impermeable polymer membrane with sequencing wells that contain nanopores and that are connected to electrical current sensing circuits to take snapshots of the electrical state of the well at a fixed sampling rate (currently 4 or 5 kHz). (B) An electrical potential across the membrane ensures an ionic flow through the nanopores. When a single‐nucleotide strand passes through the nanopore at a controlled translocation speed, this results in a characteristic disruption of the ionic current that can be basecalled into genomic information such as nucleotide sequence composition and epigenetic modifications. As nucleotides would naturally move through the nanopores too fast for the electrical circuits to detect differences in the ionic current due to individual bases, a helicase is added to the nucleotide strands as an adapter protein. This helicase docks to the nanopores, unwinds the double‐stranded DNA if applicable, and ratchets a single‐nucleotide strand through the nanopore at a controlled translocation speed. While the standard speed has been set to 400 b/s, temperature control can marginally impact translocation speed and, with that, sequencing accuracy. At a sampling rate of 4 kHz, each base is therefore assessed by approximately 10 electrical snapshots.
