Table 1.
Structure and Affinity at hP2Y 14 R of Derivatives of Potent Antagonist 1 i
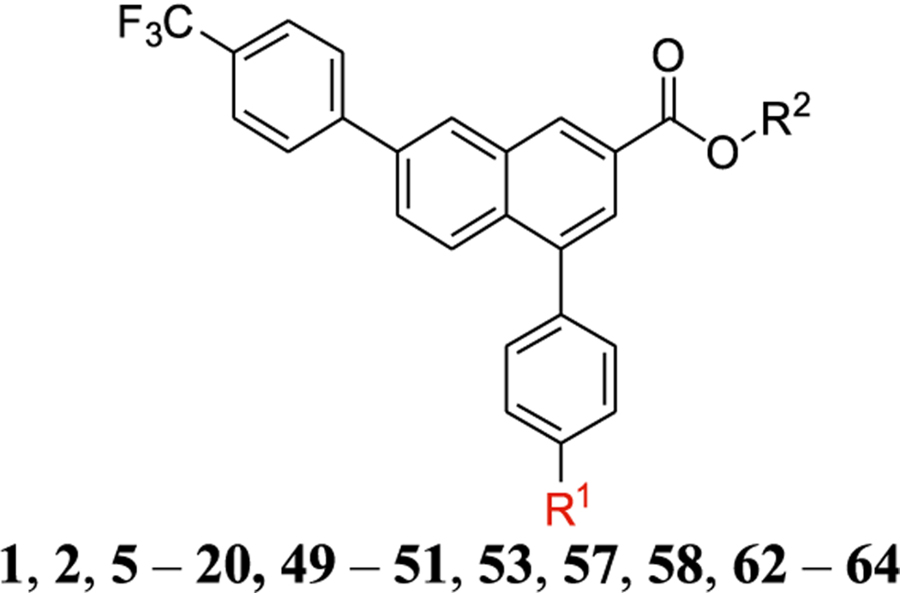
| ||||||
|---|---|---|---|---|---|---|
| No. | Structure, R1 = | IC50 (nM) | Ki (nM)d | Log D7.4e | cLogDf | log([brain]: [blood]f |
| 4-membered rings | ||||||
| 5 |
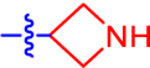
|
65.9±19.3 | 38.3 | 1.4±0.0 | 2.7 | −0.869 |
| 6 |
|
46.6±7.1 | 27.1 | 1.9±0.0 | 3.1 | −0.782 |
| 7 |
|
9.69±3.56 | 5.63 | 1.9±0.0 | 2.9 | −0.717 |
| 8 |

|
20.8±5.6 | 12.1 | 0.81±0.0 | 2.2 | −0.954 |
| 9 b |
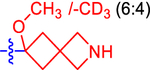
|
46.5±10.0 | 27.0 | 1.8±0.0 | 2.7 | −0.835 |
| 5-membered rings | ||||||
| 10 a |
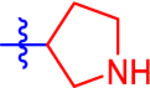
|
31.5±4.2 | 18.3 | 1.6±0.0 | 2.8 | −0.869 |
| 11 a |

|
9.48±1.66 | 5.51 | 1.1±0.0 | 2.4 | −1.08 |
| 12 a |
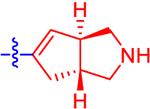
|
16.6±3.2 | 9.65 | 2.2±0.0 | 3.1 | −0.643 |
| 13 a |
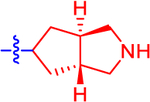
|
32.0±11.2 | 18.6 | 2.4±0.0 | 3.1 | −0.877 |
| 6-membered rings | ||||||
| 1 c |

|
7.96±0.35 | 4.63 | 1.7±0.0 | 2.9 | −0.867 |
| 2a c |
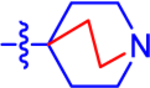
|
20.0±4.4 | 11.6 | 4.2±0.95 | 3.5 | −0.577 |
| 2b c |
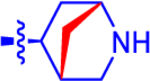
|
3.11±0.62 | 1.81 | 1.9±0.15 | 2.9 | −0.874 |
| 14 |
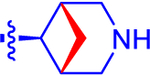
|
524±104 | 305 | 2.8±0.0 | 3.1 | −0.878 |
| 15 |
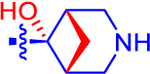
|
5.92±0.55 | 3.44 | 1.2±0.0 | 2.4 | −1.08 |
| 7-membered rings | ||||||
| 16 a |
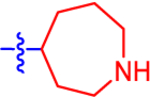
|
9.58±5.14 | 5.57 | 2.1±0.0 | 3.0 | −0.861 |
| 17 b |
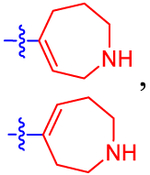
|
11.9±3.6 | 6.92 | 2.2±0.0 | 2.9 | −0.612 |
| 8-membered rings | ||||||
| 18 |
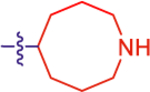
|
18.5±4.8 | 10.8 | 2.5±0.0 | 3.1 | −0.854 |
| 19 |
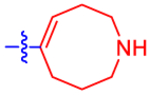
|
5 8.4± 16.4 | 34.0 | 2.9±0.0 | 3.0 | −0.616 |
| 20 |
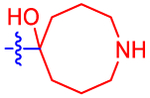
|
1780±440 | 1030 | ND | 2.2 | −1.06 |
| prodrug derivatives of 1 and IS | ||||||
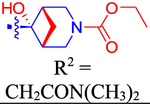
| 49 |
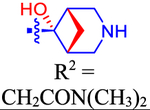
|
4810±1160 | 2800 | ND | 3.3 | 0.291 |
| 50 |
|
ND | ND | ND | 5.4 | −0.953 |
| 51 |
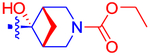
|
81.6±4.9 | 47.4 | ND | 3.3 | −1.24 |
| 62 |
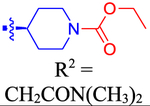
|
ND | ND | ND | 6.4 | −0.595 |
| 63 c |
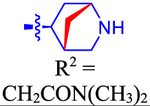
|
ND | ND | ND | 4.0 | 0.502 |
| 64 c |

|
ND | ND | ND | 6.5 | −0.664 |
| fluorescent derivative 53 and precursors | ||||||
| 53 | See Figure 1 | 25,900h | 9240h | ND | 3.6 | −1.46 |
| 57 | See Scheme S2 | 424±82 | 247 | ND | 4.3 | −1.06 |
| 58 | See Scheme S2 | 116±43g | 67.4 | ND | 3.7 | −1.08 |
Racemate.
Mixture.
Calculated from IC50 values by dividing by 1.72 (see Figure S1, Supporting Information).
Distribution coefficient Log D7.4 calculated from high-performance liquid chromatography (HPLC) retention factors k of ligands using the calibration curve of Log D7.4 vs k generated from five standards with reported log D7.4. (HPLC column: Agilent Eclipse XDB-C18 (4.6 mm × 250 mm, 5 μm); Mobile phase: A: acetonitrile, B: 10 mM TEAA, 10%−100% A in 20 min; Flow rate: 1 mL/min).
cLog D and blood–brain barrier (BBB) ratio calculated using the StarDrop program (v. 7.3.2, https://www.optibrium.com/stardrop-installers).25
Fluorescent binding IC50 value for long-chain amino derivative 58 was reported previously as 133 ± 13 nM.26
n = 1.
The affinity was determined using a whole-cell fluorescent binding assay (fluorescent tracer 52).5,17 R2 = H, and n = 3–4, unless noted. The piperidine moiety of 1 is shown in blue, while the various bridging groups, added functional groups, and other ali(hetero)cyclic rings are shown in red. ND, not determined.
