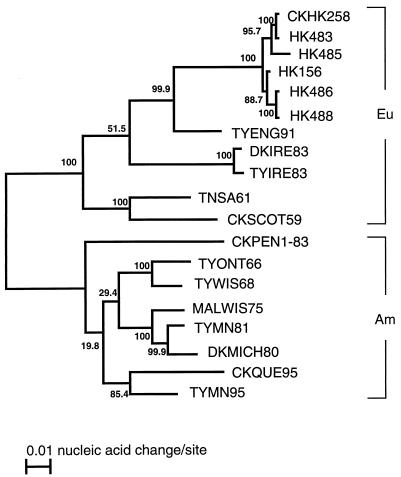
FIG. 2.
Phylogenetic relationships of the Hong Kong H5N1 virus HA genes. The nucleotide sequences of the entire HA genes were analyzed with the Clustal W program (23), which uses the algorithm of Myers and Miller (16). Numbers at nodes represent bootstrap values as a percentage of 1,000 resamplings of the data set. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes and HA sequences. Vertical lines are for spacing branches and labels; their lengths are not important. Strain abbreviations are explained elsewhere (5). Eu, Eurasian lineage; Am, American lineage.