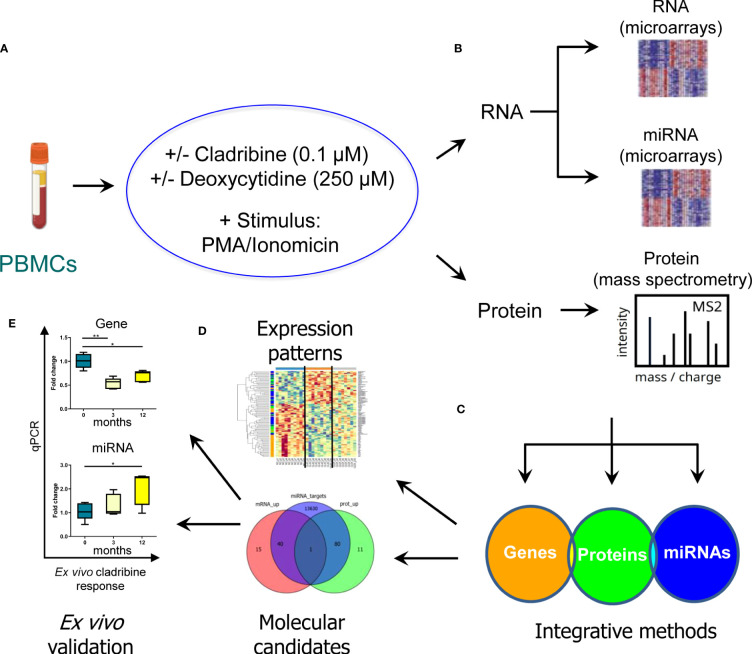
Figure 1.
Schematic representation of the strategy used to characterize the molecular effect of cladribine. In order to evaluate the molecular profiling associated with the in vitro effect of cladribine, PBMCs obtained from MS patients were first stimulated with PMA/ionomicin and then treated with cladribine in the presence or absence of an excess of deoxycytidine, a competitive substrate for DCK-dependent phosphorylation (A). Subsequently, the transcriptomic and proteomic profile was determined by means of gene and miRNA expression microarrays, and mass spectrometry in the different experimental conditions (B). Aiming to identify a multiomics biomarker signature associated with cladribine treatment, we applied two different bioinformatics approaches to integrate the three generated databases (gene, miRNAs, and proteins) (C). These analyses allowed us, on the one hand, to characterize a multiomics expression pattern associated with each experimental condition and, on the other hand, to identify potential candidate biomarkers associated with cladribine exposure (D). Finally, individual candidates were validated ex vivo in MS patients receiving treatment with cladribine (E). *p-value <0.05, **p-value <0.01.