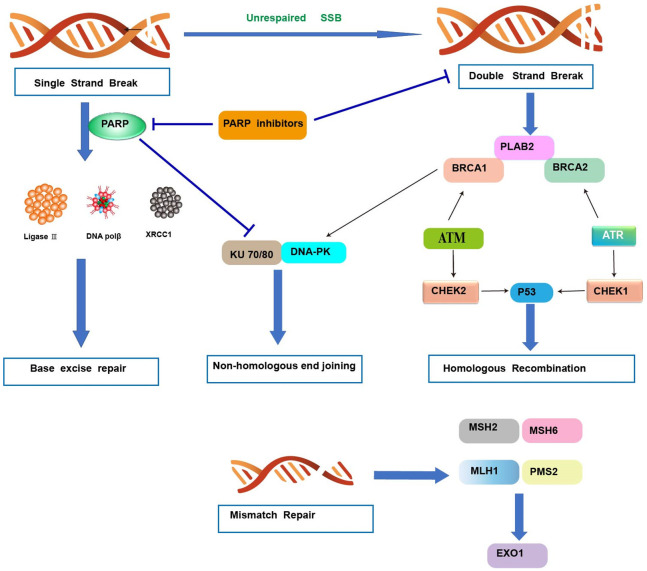
Figure 3.
DNA damage repair mechanisms. Single-strand break (SSB) repair: PARP1 detects single-strand DNA breaks and facilitates the formation of a negatively charged, branched polymer that recruits XRCC1, Ligase 3, and DNA POL β to the site of damage for ligation and repair. Inhibition of PARP1 at this stage leads to an accumulation of SSBs that ultimately results in DSBs. Double-strand break (DSB) repair: nonhomologous end joining-DNA ends are bound by Ku proteins, which are stabilized by PARP1, and form the DNA-PK (DNA-dependent protein kinase) complex following recruitment of DNA-PKcs (catalytic subunit of DNA-PK). XCCR4 and Artemis are recruited, which stabilize and recruit other repair factors to the site of damage. Homologous recombination: damage is detected by the MRN complex, which recruits and activates ATM. ATM can activate the PALB2, BRCA1, and BRCA2 complex, ATR, or CHEK2, depending on cell cycle phase. RAD51 is activated and conducts a search for a homologous template used for repair, which activates other factors necessary for repair. Ultimately, P53 is stabilized by either CHEK1 or CHEK2 and proofs the repair. Mismatch repair: a MSH2/MSH6 heterodimer recognizes and localizes mismatched base pair errors and forms a complex with MLH1 and PMS2. PARP, poly (ADP-ribose) polymerase; PARPi, PARP inhibitor; DNA polβ/δ/ε, DNA polymerase beta/delta/epsilon; XRCC1, X-ray repair cross-complementing protein 1; DNA-PKcs, DNA-dependent protein kinase catalytic subunit; KU 70/80, a.k.a XRCC6/5 (X-ray repair cross-complementing protein 6/5); ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related.