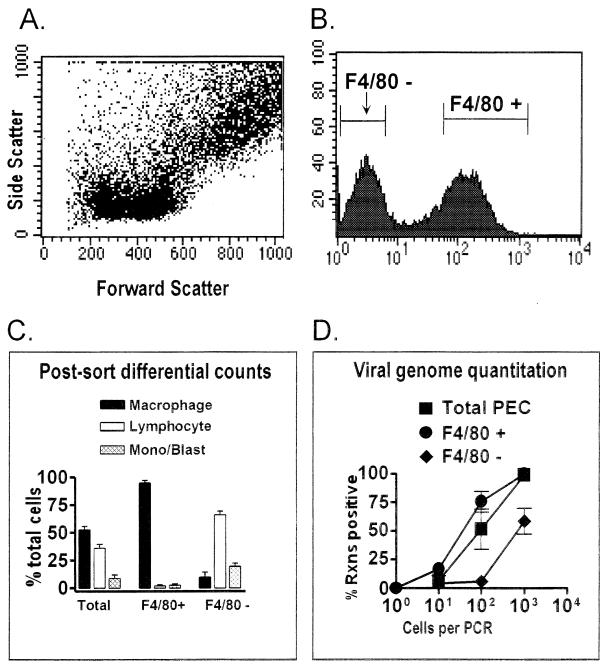
FIG. 4.
F4/80-positive peritoneal macrophages from latently infected C57BL/6 mice harbor the γHV68 genome. PECs collected from C57BL/6 mice 9 to 15 days postinfection with γHV68 were stained with F4/80. The F4/80-negative and F4/80-positive cell populations were separated by FACS sorting, and the frequency of cells carrying the γHV68 genome was quantitated by PCR. The results shown are averages of four separate experiments. In three experiments, cells were sorted as shown. In one experiment, cells were pregated into lymphocyte-enriched or macrophage-enriched populations prior to F4/80 sorting, as shown in Fig. 5 and 6. Data from the four experiments were comparable. (A) Dot plot showing forward and side scatter characteristics of peritoneal cells from a representative experiment. (B) Results of F4/80 staining and gates used for FACS sorting of F4/80-negative and F4/80-positive cell populations from a representative experiment. Cell counts are shown on the y axis, and mean fluorescence intensity is shown on the x axis. Gates for sorting were drawn tightly to prevent contamination of sorted populations. For the four experiments performed, 39 to 42% of the PECs were sorted as F4/80 negative and 39 to 44% of the PECs were sorted as F4/80 positive. (C) Pre- and postsorting differential analysis of Wright’s-stained cells from total PECs and F4/80-positive and F4/80-negative PECs. Presorting and postsorting populations were categorized by morphological criteria as macrophages, lymphocytes, or monocytes-lymphoblasts (Mono/Blast). Based on morphological criteria, monocytes could not always be distinguished from lymphoblasts. (D) Limiting-dilution quantitation of the frequency of γHV68 genome-positive cells by using total PECs and F4/80-positive and F4/80-negative PEC populations. Tenfold dilutions of each cell population were tested for the presence of the γHV68 genome by nested PCR as described in Materials and Methods. The data in panels C and D are averages of four experiments. The error bars represent the standard error of the mean. Rxns, reactions.