Fig. 1. Organizational framework for NDD pathogenesis and levels of analysis.
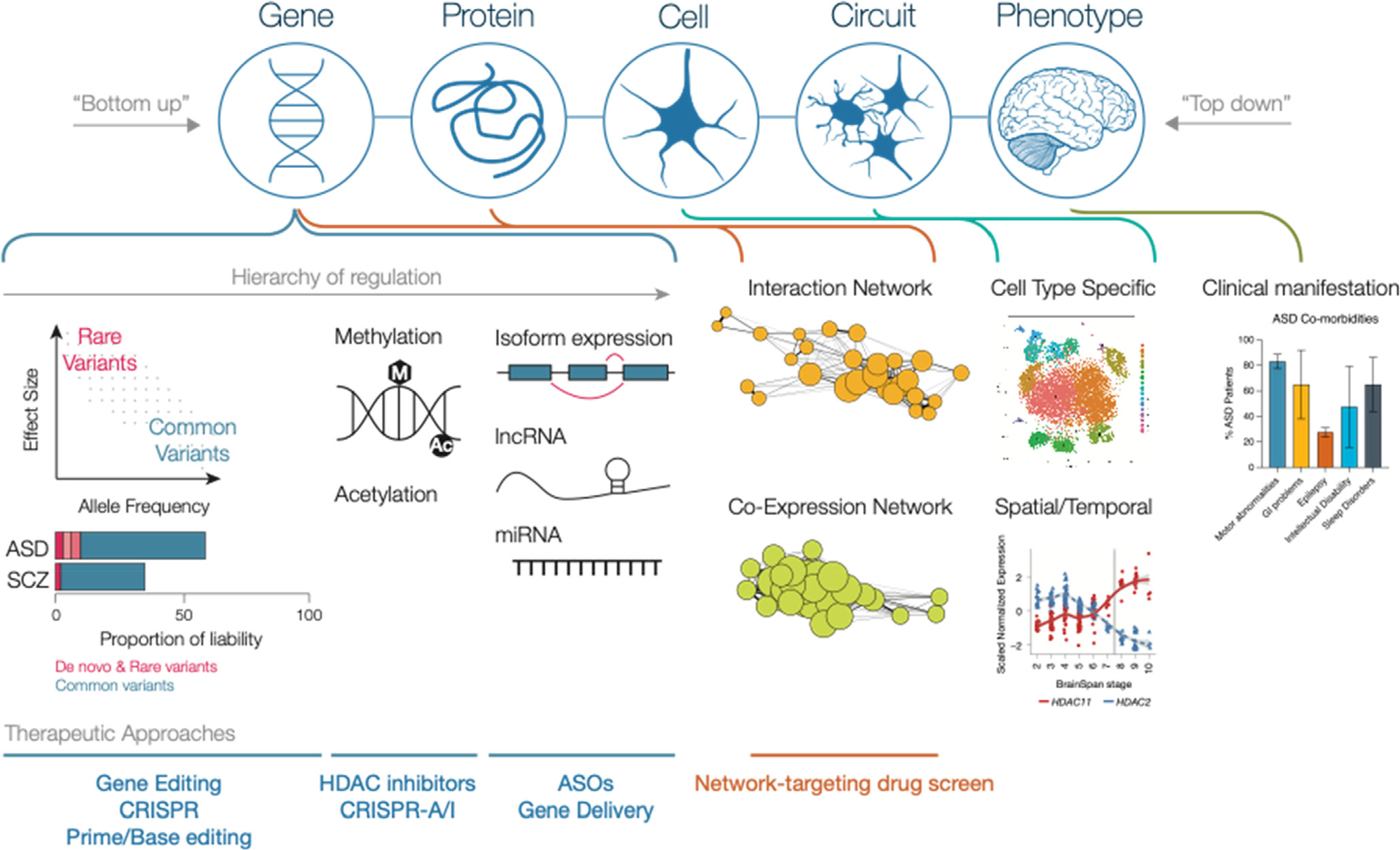
At the gene level, multiple layers of gene expression regulation have been shown to be relevant in NDD progression. Common variants underlie most of the disease risk but individually have low effect; rare mutations elicit more severe effects [44,22]. Additionally, methylation and acetylation have both been shown to regulate expression of ASD related genes. At the transcript level, additional forms of regulation, including alternative splicing, lncRNA, and miRNA modulate gene expression. The expression and interaction of genes and proteins can be explored using interaction networks (eg. PPI) and co-expression networks (eg. WGCNA) to identify biological processes and key hub genes that may prove to be attractive clinical targets. At a cell and circuit level, single cell analysis of human fetal brain [72] illustrate distinct cell types present in the developing brain. Furthermore, analyses conducted across time and between different brain regions reveal spatial and temporal differences in gene expression – for instance, a switch between expression of HDAC11 and HDAC2 in the developing prefrontal cortex as shown in the Brainspan transcriptomic data; the grey line denotes the switch from prenatal to postnatal periods [29]. In the clinic, ASD patients present with a number of co-morbidities; these may be useful as biomarkers for drug development as those with a defined phenotype can be used to screen for drug efficacy [4,8]. Therapeutic approaches can be organized within the framework by target; while gene editing approaches target individual genes or variants, network-based drug screens can link gene co-expression networks to cellular processes and circuits.
