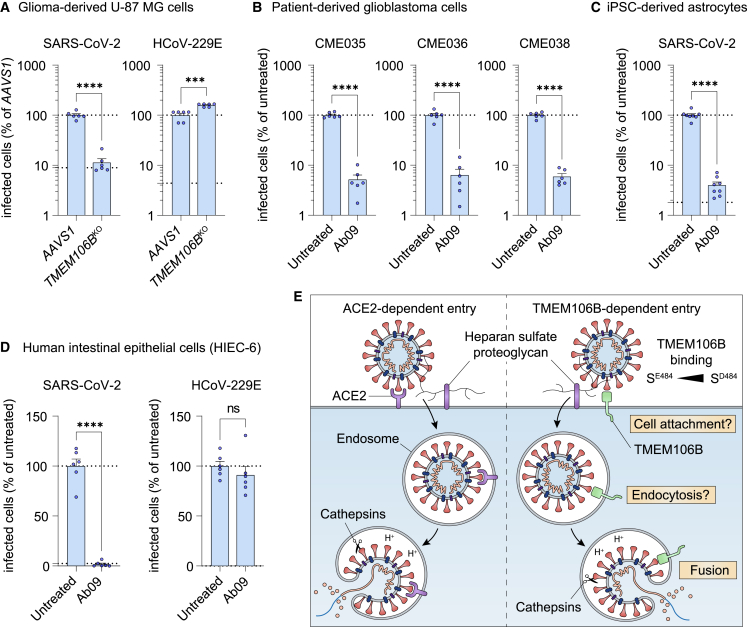
Figure 5.
Cells from various organs support SARS-CoV-2 infection via TMEM106B
(A) U-87 MG cells transduced with sgRNAs targeting TMEM106B or safe harbor locus AAVS1, infected with SARS-CoV-2 Belgium/GHB-03021/2020 or HCoV-229E, fixed after 24 (SARS-CoV-2) or 96 h (HCoV-229E).
(B) Patient-derived glioblastoma cells infected with SARS-CoV-2 Belgium/GHB-03021/2020 in the presence of anti-TMEM106B (Ab09), fixed after 24 h.
(C) iPSC-derived astrocytes infected with SARS-CoV-2 Belgium/GHB-03021/2020 in the presence of Ab09, fixed after 48 h.
(D) HIEC-6 cells infected with SARS-CoV-2 Belgium/GHB-03021/2020 or HCoV-229E in the presence of Ab09, fixed after 96 (SARS-CoV-2) or 72 h (HCoV-229E).
(A–D) Cells were stained for nucleocapsid (SARS-CoV-2) or dsRNA (HCoV-229E). Data were analyzed using two-sided unpaired t test with Welch’s correction (n = 6 wells—A, B, and D—or n = 8 wells—C —from two experiments). Upper dotted line: untreated level. Lower dotted line: detection limit. Data are mean ± SEM. ∗∗∗∗p < 0.0001; ∗∗∗0.0001 < p < 0.001; ∗∗0.001 < p < 0.01; ∗0.01 < p < 0.05; ns, not significant.
(E) Hypothetical model summarizing the two SARS-CoV-2 infection mechanisms characterized here. See also Figure S5.