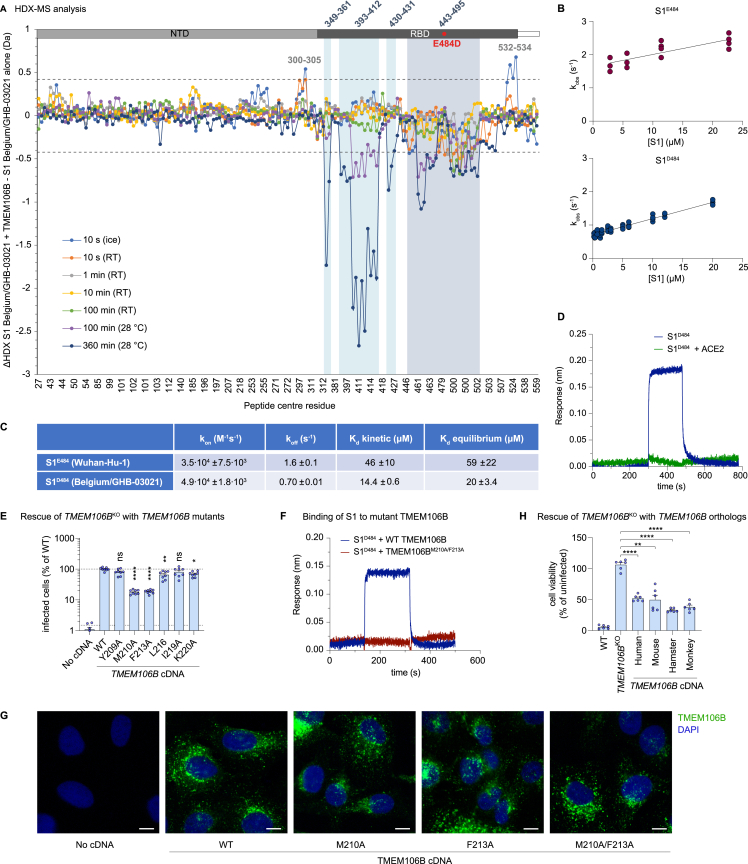
Figure S3.
Spike residue D484 and TMEM106B residues M210 and F213 enhance spike-TMEM106B binding, related to Figure 3
(A) Differences in hydrogen-deuterium exchange (ΔHDX) between the spike S1 subunit of the Belgium/GHB-03021 isolate alone and when in the presence of excess TMEM106B. Negative values indicate protection and positive values deprotection from exchange in the presence of TMEM106B. The threshold of significance calculated with 98% CI at ±0.42 Da is indicated with a dashed gray line. Peptides are arranged from the N to C terminus according to their peptide center residue. See Table S3 and Data S1 for HDX data tables and deuterium uptake plots.
(B) Biolayer interferometry results of S1 binding to immobilized TMEM106B. Data are represented as the dependence of the observed rate constant on S1 concentration for S1E484 (Wuhan-Hu-1) and S1D484 (Belgium/GHB-03021).
(C) Thermodynamic parameters for S1E484 and S1D484 subunit binding to immobilized TMEM106B. For each variant, kon was determined from the slope of the plot of kobs against (S1) for the association phase, koff was obtained from the intercept of the plot of kobs against (S1) for the association phase, KD kinetic was calculated as koff/kon, and KD equilibrium was calculated from the plot of the amplitude versus (S1).
(D) Biolayer interferometry traces of 6 μM S1D484 (blue) and 6 μM S1D484 premixed with 8 μM ACE2 (green) binding to immobilized TMEM106B (luminal domain).
(E) NCI-H1975 monoclonal TMEM106B knockout cells transduced with cDNA encoding wild-type (WT) TMEM106B or TMEM106B containing single amino acid changes, infected with SARS-CoV-2 Belgium/GHB-03021/2020. Cells were stained for dsRNA after 24 h. Infected cells were quantified by high-content imaging analysis (n = 8 wells examined over two independent experiments). Data were log-transformed and analyzed using one-way ANOVA with Dunnett’s multiple comparison test, comparing each condition with WT TMEM106B.
(F) Biolayer interferometry traces of 13.5 μM S1D484 binding to immobilized wild-type TMEM106B (blue) or mutant TMEM106BM210A/F213A (red).
(G) NCI-H1975 monoclonal TMEM106B knockout cells transduced with cDNA encoding WT or mutant TMEM106B and stained with anti-TMEM106B (Ab09) and DAPI. Representative images are shown, scale bars, 10 μm.
(H) WT or TMEM106BKO NCI-H1975 cells transduced with cDNA encoding human, mouse (mus musculus), hamster (Mesocricetus auratus), or African green monkey (Chlorocebus sabaeus) TMEM106B and infected with SARS-CoV-2 Belgium/GHB-03021/2020. Cell viability was determined by MTS assay after 3 days (n = 6 wells examined over two independent experiments). Data were analyzed using one-way ANOVA with Dunnett’s multiple comparison test, comparing each condition with TMEM106BKO cells.
(E and G) Data are mean ± SEM. ∗∗∗∗p < 0.0001; ∗∗0.001 < p < 0.01; ∗0.01 < p < 0.05; ns, not significant.