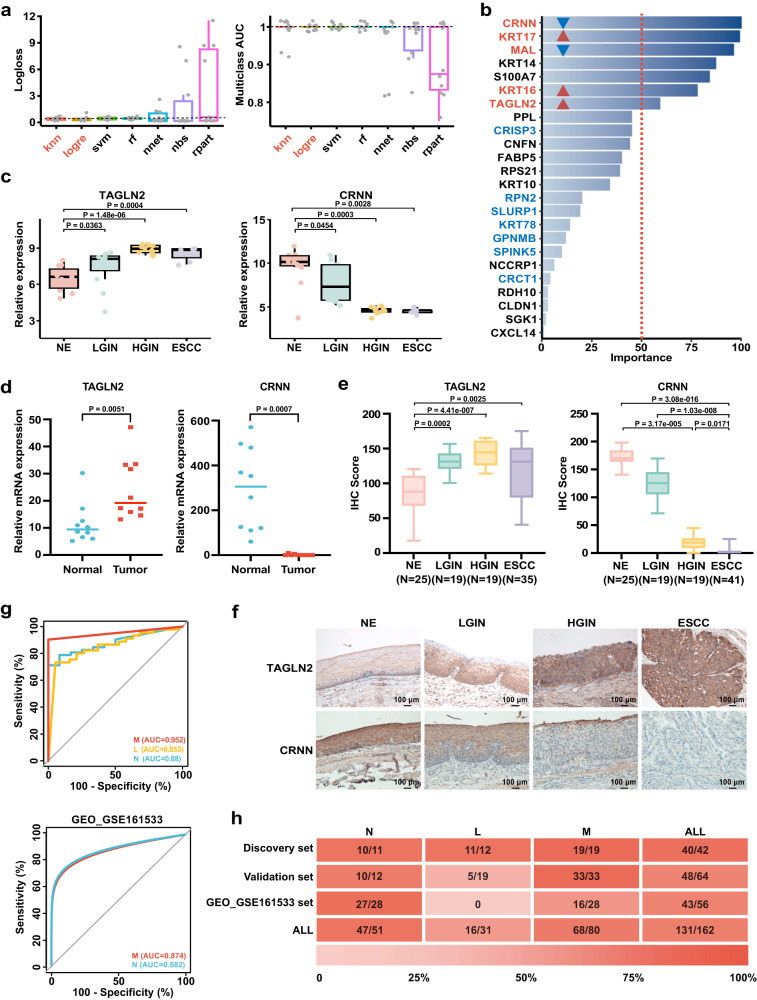
Fig. 5. Potential indicators screening for the diagnosis of ESPL and predicting the risk of ESCC.
a Logloss and multiclass AUC were analyzed by seven machine learning models (n = 10 for NE, LGIN, HGIN, and ESCC group, respectively). b The importance rank of signatures. The overlap genes between logre and knn algorithm results are marked with red and blue colors. Red color-marked genes indicated the overlap genes with importance rank >50. The red and blue triangles indicated gene expression up-regulated and down-regulated, respectively. knn (k-nearest neighbor); logre (logistic regression); svm (support vector machine); rf (random forest); nnet (neural network regression); nbs (naive bayes); rpart (decision trees). c Candidate gene (TAGLN2 and CRNN) expression in the four pathological stages by spatial WTA analysis. NE, LGIN, HGIN, and ESCC ROIs number (n = 11, 12, 12, 7). Mann–Whitney U test was used for the comparison with the NE group. In the box plots (a, c), the boxplot shows the median (central line), upper and lower quartiles (box limits), and 1.5 × interquartile range (whiskers). d Relative mRNA expression of candidate genes in paired normal and ESCC tissue. NE (n = 10), ESCC tumor tissue (n = 10) biologically independent samples. The data was analyzed by a two-tailed paired t-test. e Positive staining statistics of TAGLN2 and CRNN in the four pathological stages by IHC staining. The sample size is labeled in the figure. Kruskal–Wallis test and corrected by Dunn’s test for multiple comparisons. The boxplot shows the median (central line), upper and lower quartiles (box limits), and min to max range (whiskers). f Representative pictures of IHC stained slides (Scale bar, 100 μm, representative of n = 3 independent experiments.). g ROC plots of gene panel (TAGLN2, KRT16, CRNN) in distinguishing N and M groups from IHC staining data and GEO dataset (GSE161533). h Overall performance metrics of the prediction model for N, L, and M groups. Graduated colors indicate accuracy levels. The number in each box indicate correctly identified samples/total sample number. Source data are provided as a Source Data file.