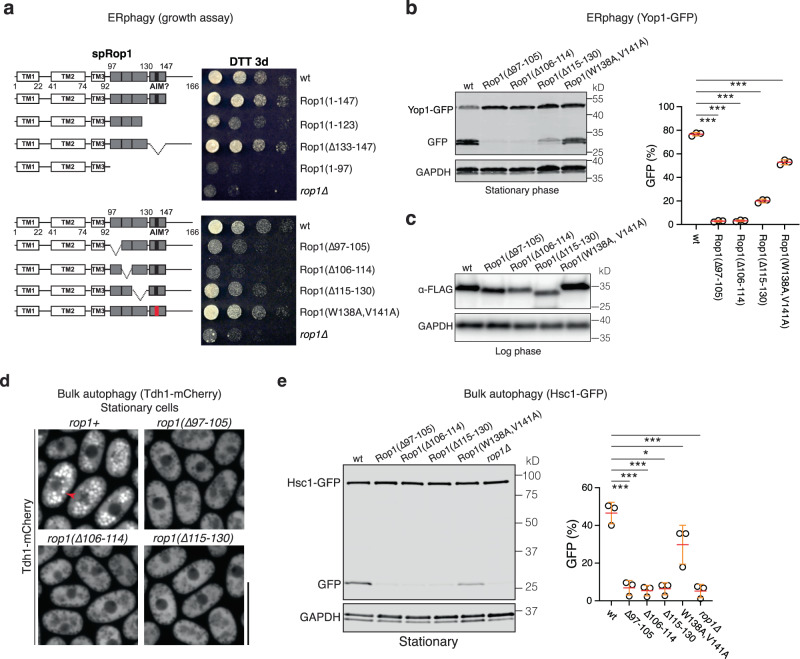
Fig. 5. Rop1’s function in autophagy requires high membrane curvature generation.
a Wild-type (wt) S. pombe Rop1 (spRop1) or the indicated mutants were expressed from the endogenous genomic locus. The cells were treated with DTT for three days (DTT 3d) and plated after serial dilution. The schemes show the domains of spRop1, with the APHs and putative AIM motif in gray and black, respectively, and deleted regions indicated. The W138A, V141A mutant is designed to abrogate the AIM motif. b ERphagy was tested by following the cleavage of a GFP-fusion of Yop1 (Yop1-GFP) in wt cells or cells expressing the indicated Rop1 mutants. Lysates from cells at stationary phase were analyzed by SDS-PAGE and immunoblotting with GFP antibodies. Blotting with GAPDH antibodies served as loading control. The right panel shows quantification of three independent experiments (means and SD). *** indicates significant differences with p-values < 0.001, calculated from two-tailed Student’s t tests. The exact p-values are listed in the Source Data file. c wt or mutant Rop1 used in (b) were FLAG-tagged. Lysates from logarithmically growing cells were analyzed by immunoblotting for FLAG. d Bulk autophagy was tested by following the relocalization of a mCherry fusion of the cytosolic protein Tdh1 (Tdh1-mCh) expressed at endogenous levels in wt cells or cells expressing the indicated Rop1 mutants. Stationary phase cells were analyzed by fluorescence microscopy. Scale bar, 10 µm. e Bulk autophagy was followed by the vacuolar cleavage of a GFP-fusion of the cytosolic protein Hsc1 (Hsc1-GFP) in the indicated strains grown to stationary phase. The right panel shows the percentage of Hsc1-GFP cleavage (means and SD). *p-value 0.07; *** significant differences with p-values < 0.001, calculated from two-tailed Student’s t tests. The exact p-values are listed in the Source Data file.