FIGURE 1.
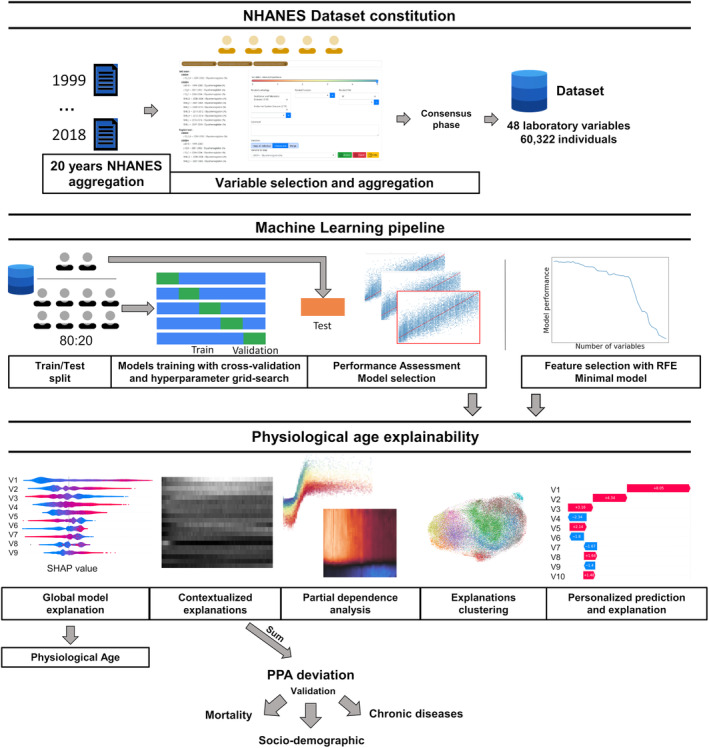
Machine learning analysis pipeline. All data from the National Health and Nutrition Examination Surveys (i.e., NHANES study) 1999–2018 were collected. A large, consistent database containing the maximal number of common biological variables reported on the maximal number of subjects, and with minimal missing data was generated. This resulted in a dataset with 60,402 individuals with 48 biological variables and 0.01% missing data. Using this dataset, five classes of algorithm models were trained, tested and compared based on performance. The XGBoost model with custom loss was considered (see Figure 2), and explainability was computed using SHAP values for the personalized physiological age (PPA) estimation. Deviation of PPA from chronological age is therefore the sum of the contextualized SHAP contributions of all the laboratory variables for a given subject (PPA deviation). Partial dependence plots and heatmaps of SHAP values also identify the precise range of biological values and thresholds for each variable and age group delineating accelerated or reduced aging. Clustering of SHAP values identifies specific PPA profiles. Finally, using recursive feature elimination, the list of variables was reduced to 26 biological variables without significant loss of model performance, providing a ready‐to‐use personalized and explainable model that is potentially clinically useful for monitoring physiological age to achieve healthy aging. PPA deviation was validated as a predictor of lifespan but also a risk factor for chronic diseases.
