Abstract
Keloids are benign, fibroproliferative dermal tumors that typically form owing to abnormal wound healing. The current standard of care is generally ineffective and does not prevent recurrence. To characterize keloid scars and better understand the mechanism of their formation, we performed transcriptomic profiling of keloid biopsies from a total of 25 subjects of diverse racial and ethnic origins, 15 of whom provided a paired nonlesional sample, a longitudinal sample, or both. The transcriptomic signature of nonlesional skin biopsies from subjects with keloids resembled that of control skin at baseline but shifted to closely match that of keloid skin after dermal trauma. Peripheral keloid skin and rebiopsied surrounding normal skin both showed upregulation of epithelial–mesenchymal transition markers, extracellular matrix organization, and collagen genes. These keloid signatures strongly overlapped those from healthy wound healing studies, usually with greater perturbations, reinforcing our understanding of keloids as dysregulated and exuberant wound healing. In addition, 219 genes uniquely regulated in keloids but not in normal injured or uninjured skin were also identified. This study provides insights into mature and developing keloid signatures that can act as a basis for further validation and target identification in the search for transformative keloid treatments.
Introduction
Keloid scars are fibroproliferative dermal tumors that represent an aberration from the normal wound healing response, culminating in the overgrowth of dense fibrous tissue that extends beyond the borders of the initial injury. Although classified as a benign dermal growth, keloids demonstrate biological features akin to malignant tumors such as hyperproliferation, apoptosis resistance, and invasion (Mari et al., 2015; Tan et al., 2019). Keloid scars can cause pain, pruritus, and contracture, leading to significant physical and psychological burden (Gauglitz et al., 2011). Despite the many available medical and surgical treatment options, outcomes remain poor, and recurrence rates remain high, underscoring the need for further elucidation of keloid pathogenesis (Betarbet and Blalock, 2020).
The exact molecular and cellular mechanisms underlying keloid pathology remain obscure, but genome-wide expression profiling studies yielded clues. Bulk-RNA sequencing comparison of lesional with nonlesional skin from African American subjects found that inflammation pathways, including Th1, Th2, Th17/22, and Jak3, were associated with lesional skin compared with association with healthy controls (Wu et al., 2020). Studies have also implicated the role of microRNAs, long noncoding RNAs, and circular RNAs in keloid development and physiology (Onoufriadis et al., 2018; Wang et al., 2019a). The role of specific cell types in the formation of keloids has also been examined, either by characterizing cells isolated from keloid tissue or in an unbiased fashion using single-cell RNA-sequencing profiling. Two separate single-cell studies reported the expansion of fibroblasts in keloid tissue, with Deng et al. (2021) additionally identifying a keloid-specific mesenchymal fibroblast subpopulation (Deng et al., 2021; Liu et al., 2022). Endothelial dysfunction in the reticular dermis is thought to cause both keloids and hypertrophic scars, with recent single-cell data supporting an increased abundance of vascular endothelial cells in keloids (Liu et al., 2022). Matsumoto et al. (2020) specifically investigated changes in the gene expression profile of vascular endothelial cells isolated from keloids, and others have characterized immune cells (including mast cells, T cells, dendritic cells, and M2 macrophages) infiltrating keloid tissue and playing a role in inflammation and fibrosis (Wu et al., 2020; Xu et al., 2020).
Comparing keloid lesions with healthy skin is crucial in our understanding of keloid development and biology. However, many of the components, including inflammatory pathways, the extracellular matrix, collagen production, TGFβ signaling, M2 macrophage enrichment, and immune cell infiltration, are also components of the normal wound healing process. To isolate the dysregulated component of the healing process in keloids and identify any keloid-specific genes, the keloid signature must be compared with a normal wound-healing signature.
In this study, we obtained a total of 60 skin-punch biopsies from patients with keloids, the largest sample cohort published to date (Table 1). We included both lesional and nonlesional skin and incorporated baseline biopsies and follow-up rebiopsies from subjects with keloids who are of diverse races and ethnicities to elucidate the signature of developing keloids for comparison with both mature keloids and healthy wound healing.
Table 1.
Study Design
| Race/Ethnicity | Sex | Lesion Body Region | Nonlesional Body Region | Baseline Biopsies | Rebiopsies/Follow-Up Visits | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| African American | F | Abdomen | Ctrl | ||||||||
| African American | F | Back | Ctrl | NonLes_b | Nonlesional baseline biopsy (n = 8) | ||||||
| Asian | M | Arm | Ctrl | NonLes_r | Nonlesional rebiopsy (n = 5) | ||||||
| White | F | Back | Ctrl | KEar | Keloid ear biopsy (n = 9) | ||||||
| White (n = 2) | F | Abdomen | Ctrl | KE_b | Keloid edge baseline biopsy (n = 25) | ||||||
| African American | F | Abdominoplasty | AbPlast | KE_r | Keloid edge rebiopsy (n = 13) | ||||||
| Hispanic/Latino (n = 2) | F | Abdominoplasty | AbPlast | Ctrl | Control (n = 6) | ||||||
| Unknown | F | Abdominoplasty | AbPlast | AbPlast | Abdominoplasty (n = 10) | ||||||
| White (n = 6) | F | Abdominoplasty | AbPlast | ||||||||
| AfricanAmerican (n = 3) | F | Ear | KEar | ||||||||
| Asian | F | Ear | KEar | ||||||||
| Hispanic/Latino | M | Ear | KEar | ||||||||
| White | F | Ear | KEar | ||||||||
| Asian (n = 2) | F | Back/ear | KE_b | KEar | |||||||
| African American | F | Arm | KE_b | ||||||||
| African American | F | Back | KE_b | ||||||||
| Asian | M | Arm | KE_b | ||||||||
| Hispanic/Latino | M | Back | KE_b | ||||||||
| NativeHawaiian/PacificIslander | F | Arm | KE_b | ||||||||
| White | M | Chest | KE_b | ||||||||
| White | M | Back | KE_b | ||||||||
| African American | M | Arm | Arm | KE_b | NonLes_b | ||||||
| African American | F | Back | Back | KE_b | NonLes_b | ||||||
| African American | M | Chest | Chest | KE_b | NonLes_b | ||||||
| African American | M | Chest | KE_b | 47d | KE_r | ||||||
| African American | M | Arm | KE_b | 49d | KE_r | ||||||
| White | F | Arm | KE_b | 52d | KE_r | ||||||
| White | F | Chest | KE_b | 56d | KE_r | ||||||
| Asian | M | Arm | KE_b | 61d | KE_r | ||||||
| African American | F | Back | KE_b | 71d | KE_r | ||||||
| Asian | M | Chest | KE_b | 93d | KE_r | ||||||
| African American | M | Chest | KE_b | 97d | KE_r | ||||||
| AfricanAmerican | M | Chest | Arm | KE_b | NonLes_b | 62d | KE_r | NonLes_r | |||
| AfricanAmerican | M | Back | Back | KE_b | NonLes_b | 90d | KE_r | NonLes_r | |||
| AfricanAmerican | F | Chest | Chest | KE_b | NonLes_b | 90d | KE_r | NonLes_r | |||
| AfricanAmerican | F | Chest | Chest | KE_b | NonLes_b | 98d | KE_r | NonLes_r | |||
| African American | F | Abdomen | Abdomen | KE_b | NonLes_b | 69d | KEar | 107d | KE_r | NonLes_r | |
Abbreviations: Ctrl, control; F, female; M, male.
The table shows the study participants and the biopsies collected. The number of subjects with identical information is indicated inside red parenthesis. Baseline lesions, taken from the periphery without adjacent normal skin and untreated for a minimum of 6 months, were obtained from the ear (KEar) or torso (KE_b) (broadly categorized into arm, chest, abdomen, and back). Biopsy locations were determined by subject choice. Nonlesional biopsies (NonLes_b) were obtained from within 5 cm of lesional biopsy (generally within ∼2–3 cm). Rebiopsies (KE_r and NonLes_r) were taken from the same location as baseline biopsies, with days from baseline biopsy indicated in gray. Two Ctrl groups consisted of healthy volunteers (Ctrl) and abdominoplasty discard skin (AbPlast). Intralesional triamcinolone injections were given if the subject developed keloid at the site of nonlesional biopsy.
Results
Mature keloid signature
Mature keloids at baseline biopsy (i.e., KE_baseline, n = 25) showed profound transcriptomic changes compared with control skin (i.e., control, n = 6), with 4,670 differentially expressed genes perturbed by at least 50% (fold change > 1.5). The majority (64%) of differentially expressed genes were upregulated. This mature keloid signature was similarly perturbed in keloids at rebiopsy (i.e., KE_rebiopsy, n = 13) and ear keloids at baseline (i.e., KEar, n = 9) but not in nonlesional skin at baseline (i.e., NonLes_baseline, n = 8) or abdominoplasty tissue from nonkeloid subjects (I.e., AbPlast, n = 10). Nonlesional skin from subjects with keloid at rebiopsy (i.e., NonLes_rebiopsy, n = 5) also mirrored this signature, suggesting that the initial biopsy triggered molecular changes associated with keloid formation (Figure 1). The strength of this signature was variable across keloid samples but was more pronounced after rebiopsy than at baseline (Figure 2a).
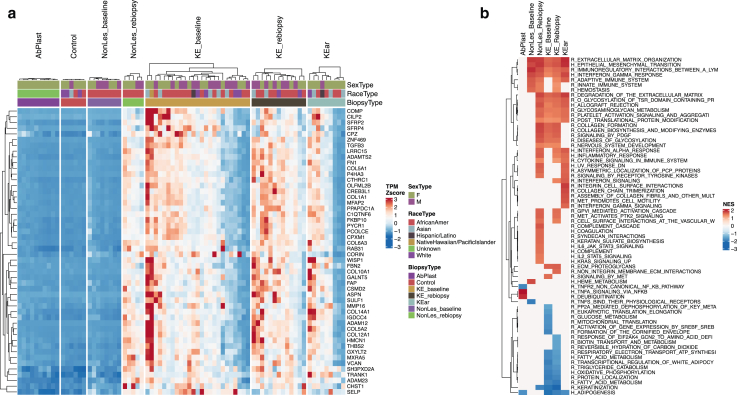
Figure 1.
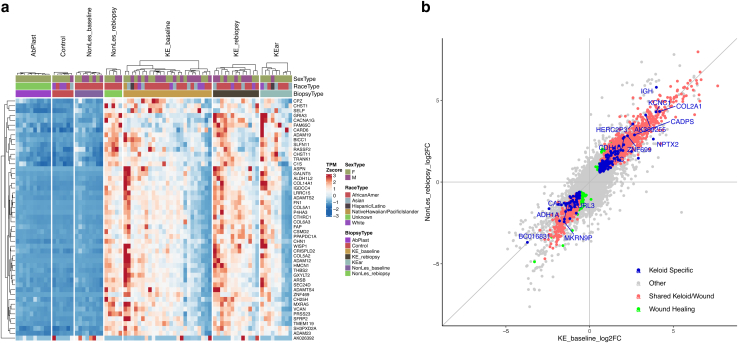
Cell-type markers heatmap. A Heatmap of TPM Z scores (percentile capped TPM at 5th and 95th) for cell-type marker genes is presented. Cell types included are skin cells (fibroblasts, keratinocytes, neural cells, melanocytes, endothelial) and immune cells (macrophage, mast cells, dendritic cells, multiple types of T cells, B cells, neutrophils). Cell cycling markers, collagens, and top 10 DEGs from the keloid baseline signature were also included as references. KE_baseline signature and a subset of collagens were upregulated in keloid tissue. Fibroblast markers closely follow the same pattern. Biological sex, race, and biopsy type are indicated in the figure column header. DEG, differentially expressed gene; K, keratin; TPM, transcripts per million.
Figure 2.
Keloid signature and pathway enrichment analysis. (a) Data showing the top 50 genes in keloid signature by P-value (KE_b compared with Ctrl). Heatmap of TPM Z scores (percentile capped at 5th and 95th) show that this signature is reflected across keloid groups (KE_rebiopsy and KEar) as well as post-trauma nonlesional skin (NonLes_rebiopsy) but is absent in both healthy skin groups (AbPlast and Ctrl) and baseline nonlesional skin (NonLes_baseline). Some heterogeneity of signature intensity is observed within each keloid group. Biological sex, race, and biopsy type are indicated in the figure column header. (b) Select GSEA results, ordered by maximum absolute NES across comparison groups. Missing values indicate nonsignificance. Pathways tested came from Hallmark (denoted as H_) or Reactome (denoted as R_) databases. Repetitive and irrelevant pathways were excluded. Ctrl, control; GSEA, gene set enrichment analysis; NES, normalized enrichment score; TPM, transcripts per million.
Pathway-level analysis highlighted strong upregulation of collagen formation (Figures 2b and 3), extracellular matrix remodeling (COMP, SFRP4, FN1, VCAN, BGN, TNC), and PDGF signaling (THBS1, THBS2, SPP1, PDGFRA, PDGFRB, signal transducer and activator of transcription 3 gene STAT3), consistent with expectations for a fibroproliferative disorder. Upregulation of these pathways and fibroblast markers (COL10A1, FAP, COL1A1, COL3A1) indicate the likely proliferation of fibroblasts in keloid tissue (Figure 1).
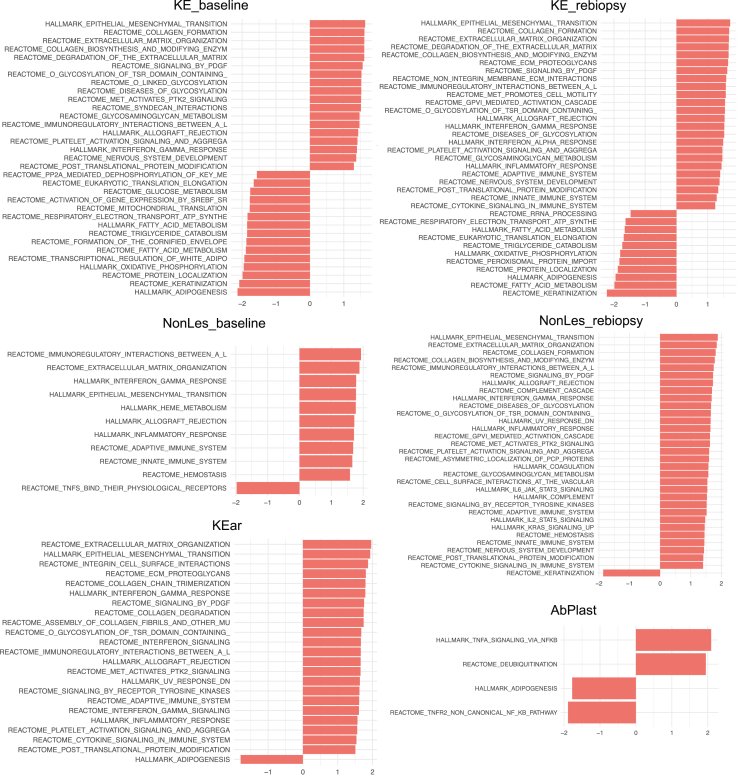
Figure 3.
GSEA results. GSEA results for each phenotype group are shown. Only significant pathways are shown. Pathways tested came from Hallmark or Reactome databases. GSEA, gene set enrichment analysis.
We investigated immune response and immune cell infiltration in keloid tissue. There was evidence of upregulation in immune response, including enrichment of the IFN-γ response pathway (IL4R, IL6, signal transducer and activator of transcription 2 gene STAT2, VCAM1) (Figures 2b and 3). Immune cells, including macrophages, mast cells, B cells, and T cells, were enriched in a subset of keloid groups (Figure 1). Interestingly, this enrichment was not necessarily correlated with the strength of enrichment in extracellular matrix and fibrosis genes, suggesting possible keloid subtypes with and without an inflammatory component.
Endothelial cells appear to be similarly enriched, with endothelial cell markers clustering with immune cell markers rather than fibroblasts (Figure 1). Finally, nervous system development is enriched in keloid groups, with leading edge genes comprising collagens, ITGA10, NRP2, SEMA3A, SLIT2, and more. We observed an enrichment of neural cell markers NRXN1 and NRXN2 mostly prominently in the KE_baseline group (Figure 1).
Cell-type composition changes in mature keloid signature
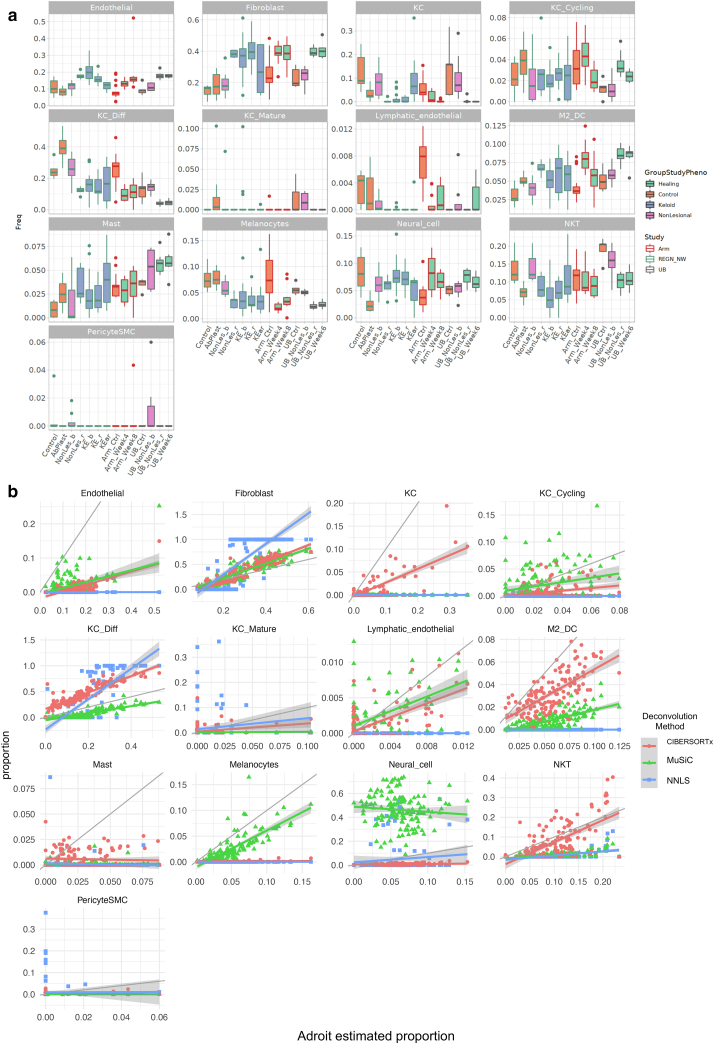
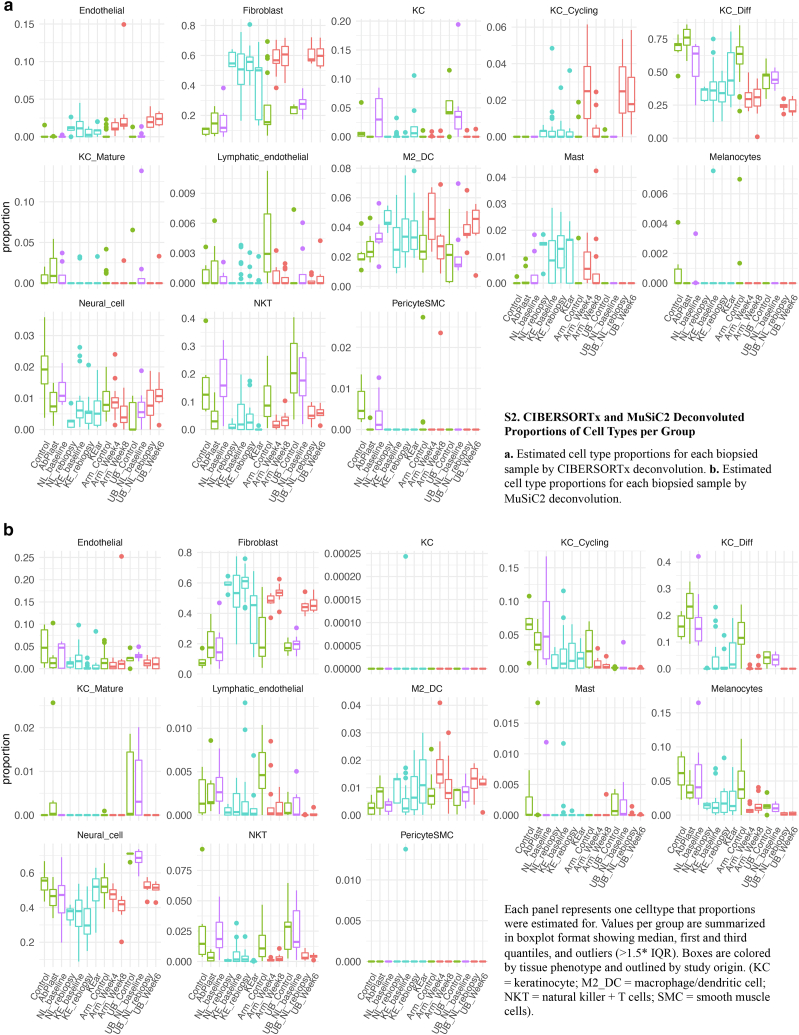
Cell-type composition information from bulk RNA was further inferred through deconvolution using cell type–specific signatures derived from a published keloid and normal scar single-cell dataset (Deng et al., 2021). Cell types from Deng et al. (2021) were reannotated for cell-type marker generation (Figure 4). Three different deconvolution algorithms were applied: AdRoit (Yang et al., 2021) (Figure 5a), CIBERSORTx (Newman et al., 2019), and MuSiC2 (Wang et al., 2019b) (Figure 6). Although absolute estimated numbers do not agree completely between algorithms, a few general trends appear consistent (Figure 5b). Estimated total fibroblast proportions show a strong correlation across all three algorithms, with strong enrichment in keloid and wound healing samples (Figure 5). Endothelial cells and M2 macrophages/dendritic cells show a similar pattern of increase. In contrast, estimated NK T-cell proportions were nearly identical between AdRoit and CIBERSORTx and show an overall reduction in keloids and wound-healing skin. Other cell types that showed a proportional reduction included multiple keratinocyte populations and possibly melanocytes.
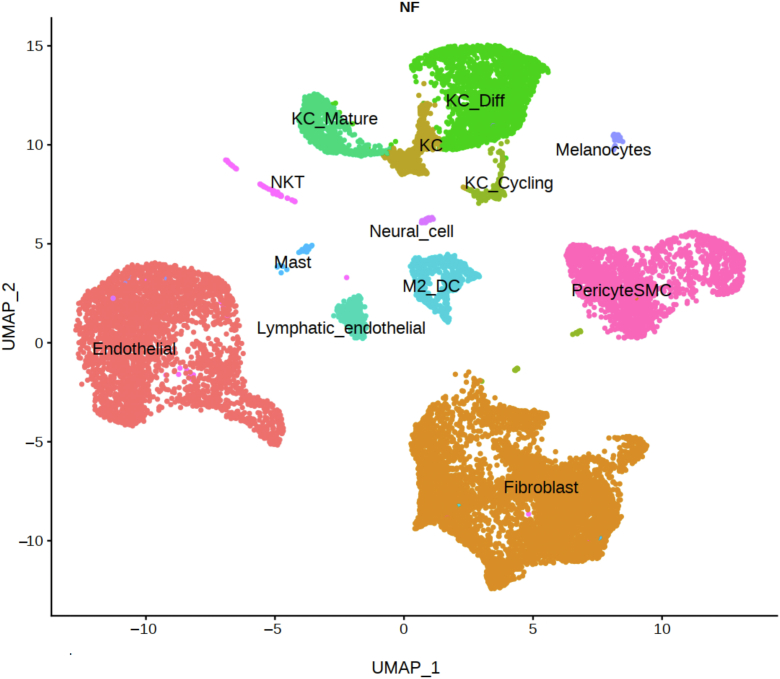
Figure 4.
UMAP of reannotated Deng et al. (2021) cell types used in bulk deconvolution. Presented is a UMAP of reanalyzed and reannotated public datasets from Deng et al. (2021). Major cell types include fibroblasts, endothelial cells, and pericyte/smooth muscle cells. Other cell types are named as KC_Diff (differentiated keratinocytes), KC_Mature (mature keratinocytes), KC_Cycling, (cycling keratinocytes), KC (keratinocytes), NKT (NK and T cells), and M2_DC (macrophage and dendritic cells). UMAP, Uniform Manifold Approximation and Projection.
Figure 5.
Deconvoluted cell-type compositions. (a) Estimated cell-type proportions for each biopsied sample by AdRoit deconvolution. Each panel represents one cell type that the proportions were estimated for. Values per group are summarized in boxplot format showing median, first and third quantiles, and outliers (>1.5∗ IQR). Boxes are colored by tissue phenotype and outlined by study origin. KC denotes keratinocyte, M2_DC denotes macrophage/dendritic cell, NKT denotes NK + T cells, and SMC denotes smooth muscle cells. (b) Correlations between cell-type proportion estimates from AdRoit (X axes), compared with those from CIBERSORTx and MuSiC2 (Y axes). Ctrl, control; IQR, interquartile range.
Figure 6.
CIBERSORTx- and MuSiC2-deconvoluted proportions of cell types per group. (a) Estimated cell-type proportions for each biopsied sample by CIBERSORTx deconvolution. (b) Estimated cell-type proportions for each biopsied sample by MuSiC2 deconvolution. Each panel represents one cell type that the proportions were estimated for. Values per group are summarized in boxplot format showing median, first and third quantiles, and outliers (>1.5∗ IQR). Boxes are colored by tissue phenotype and outlined by study origin. KC denotes keratinocyte, M2_DC denotes macrophage/dendritic cell, NKT denotes NK + T cells, and SMC denotes smooth muscle cells. IQR, interquartile range.
Mature keloid signature versus wound healing
To distinguish the mature keloid signature from that of wound healing, two public datasets (Onoufriadis et al., 2018; Ud-Din et al., 2021) profiling tissue biopsies from control skin at baseline and rebiopsied wounded skin from the same site were identified for comparison. In Ud-Din et al. (2021), we identified 17 baseline biopsies that were taken from the inner arm of control volunteers who had received no treatment or only placebos (i.e., Arm_Control) and 12 rebiopsies that were taken at week 4 (i.e., Arm_Week4) and again at week 8 (i.e., Arm_Week8) for subjects who were only treated with placebo. In Onoufriadis et al. (2018), baseline biopsies were taken from the upper buttock of six control volunteers (i.e., UB_Control), with rebiopsies taken at week 6 (i.e., UB_Week6). In addition, baseline biopsies were taken from the upper buttock of eight keloid-prone volunteers (i.e, UB_NonLes_baseline), with rebiopsies taken at week 6 (i.e., UB_NonLes_rebiopsy). Wound-healing signatures were derived for both studies by comparing the respective rebiopsies with their control baseline biopsies (i.e., Arm_Week4/Arm_Week8 against Arm_Control and UB_week6 against UB_Control) (Figure 7a).
Figure 7.
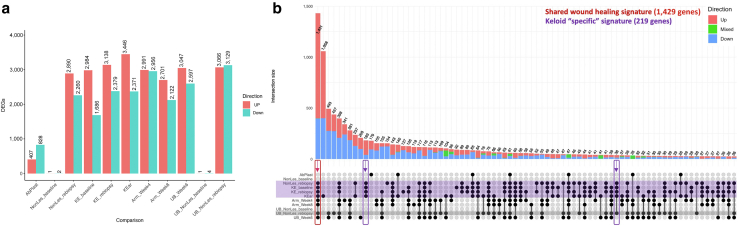
Keloid and wound-healing signature overlaps reveal keloid-associated gene sets. (a) Total number of DEGs (adjusted P-value < 0.05 and fold change > 1.5) in each group compared with those of their respective control groups, split by direction of change. (b) Upset plot summarizing the overlap of signatures in a. Upper panel bar plots: Each vertical bar shows the subset of DEGs that is significant across all the groups with filled-in black bubbles in the column directly below the bar. The proportions of this subset of genes that are upregulated across all indicated groups are represented by the red part of the bar: downregulated genes in blue and inconsistent genes in green. Lower panel: Keloid groups are highlighted in purple. Potentially developing keloid group is highlighted in gray. The red box and arrow indicate the DEG subset significant in all lesional and wound-healing groups, defined as the shared wound healing signature. Purple boxes and arrows indicate the DEG subsets only significant in keloid groups, defined as a keloid-associated signature. DEG, differentially expressed gene; F, female; M, male.
Two nonexclusive hypotheses for the keloid signature were investigated. The first hypothesis is that the keloid signature is the normal wound healing response but persistent and exacerbated. This hypothesis would be supported by a large number of genes being similarly differentially expressed in all keloid and healthy wound healing groups. The largest intersection between the signatures of the 11 comparison groups conformed to this hypothesis. This intersection contained 1,431 differentially expressed genes present exclusively in the eight keloid and wound-healing groups. Any genes that showed mixed directions of expression change across comparison groups were removed as noise. The remaining 1,429 genes were defined as a shared wound healing (SWH) signature.
The second hypothesis is that there is a keloid-associated signature (KAS) not part of the normal wound-healing response. This hypothesis predicts that there are genes differentially expressed in the keloid groups (KE_baseline, KE_rebiopsy, KEar, NonLes_rebiopsy) but not in the healthy wound-healing groups (Arm_Week4, Arm_Week8, UB_Week6). NonLes_rebiopsy was included as a keloid group because five of seven subjects developed keloids at follow-up. In contrast, the UB_NonLes_rebiopsy group was not because just two of eight subjects developed keloids. Two intersections conforming to this hypothesis were combined, with a sum of 219 genes to define the KAS. The combinations with which these two signatures were obtained are presented in an UpSet plot (Figure 7b).
To investigate whether the genes in the SWH list are differentially regulated to a more extreme degree in keloids than in wound healing, the log2 fold changes of all genes expressed in the skin were compared between KE_baseline and wound-healing signatures. There appeared to be a stronger upregulation of the SWH genes, with the largest log2 fold changes in keloids than in the inner arm and to a smaller degree in the upper buttock (Figure 8). Specifically, there were 45 SWH genes that showed stronger upregulation in KE_baseline than in wound healing groups (Arm_Week4, Arm_Week8, UB_Week6), with residual from 1:1 line in the top 2.5% of all genes across all three comparisons (Supplementary Data).
Figure 8.
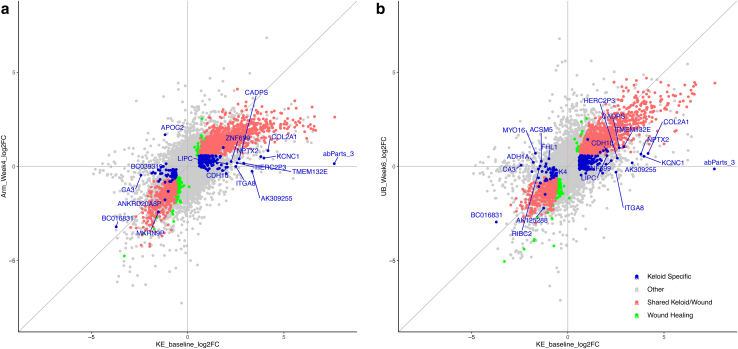
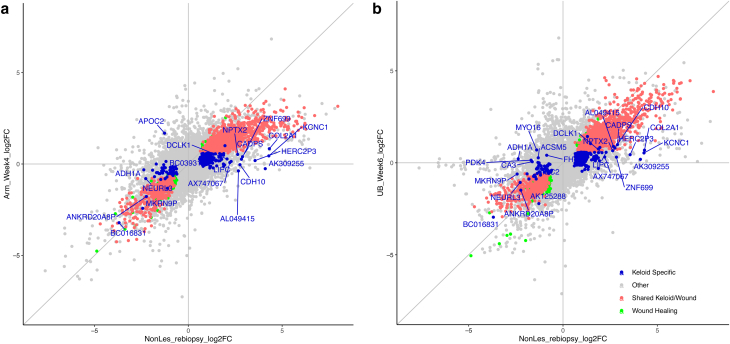
Scatterplot of SWH and KAS FCs in keloid baseline versus in healing skins. Left panel: FC of genes in lesions (KE_baseline compared with Ctrl) against FC of genes in arm wound healing (Arm_Week4 compared with Arm_Ctrl). Right panel: FC of genes in lesions (KE_baseline compared with Ctrl) against FC of genes in upper buttock wound healing (UB_Week6 compared with UB_Ctrl). All genes are represented in gray. SWH genes are colored red, and KAS genes are colored blue. Genes in green are wound-healing–specific genes not significant in any of the keloid groups. KAS genes with the largest fold changes in either direction are labeled by name. Ctrl, control; FC, fold change; KAS, keloid-associated signature; SWH, shared wound healing.
For the most part, KAS genes did not show significant divergence from the expression pattern of SWH genes, but there was a small subset of 12 KAS genes that were significantly more upregulated in keloids than in normal wound healing, including COL2A1, NPTX2, and KCNC1 (Figure 8). A pathway analysis of all 57 SWH and KAS that were significantly upregulated in keloids compared with those in normal wound healing confirmed the upregulation of pathways associated with collagen formation and extracellular matrix organization (Supplementary Data).
Trauma from skin-punch biopsy (between 47 and 107 days after baseline biopsy) resulted in a transcriptomic change in nonlesional skin similar to the mature keloid signature, with a tight correlation between the log2 fold change values of KE_Baseline and NonLes_rebiopsy (Figure 9). The KAS genes most prominent in healing nonlesional skin that went on to develop keloids are COL2A1 and KCNC1 (Figure 10).
Figure 9.
Scatterplot of SWH and KAS FC in nonlesional rebiopsy versus keloid baseline. (a) Data shown for top 50 genes in rebiopsied nonlesional skin signature by P-value (NonLes_rebiopsy compared with Ctrl). Heatmap of TPM Z scores (percentile capped at 5th and 95th) shows that this signature is reflected across keloid groups (KE_baseline, KE_rebiopsy, and KEar) but is absent in both healthy skin groups (AbPlast and Ctrl) and baseline nonlesional skin (NonLes_baseline). Some heterogeneity of signature intensity is observed within each keloid group. Biological sex, race, and biopsy type are indicated in the figure column header. (b) Scatterplots of the fold change of all genes (in gray) in KE_baseline signature against NonLes_rebiopsy. Genes in the SWH are colored red, and genes from the KAS are colored blue on the topmost layer. Genes in green are wound-healing–specific genes not significant in any of the keloid groups. Ctrl, control; F, female; FC, fold change; KAS, keloid-associated signature; M, male; SWH, shared wound healing; TPM, transcripts per million.
Figure 10.
Scatterplot of SWH and KAS FCs in nonlesional rebiopsy versus arm and UB. Left panel: FC of genes in nonlesional rebiopsies (NonLes_rebiopsy compared with Ctrl) against FC of genes in arm wound healing (Arm_Week4 compared with Arm_Ctrl). Right panel: FC of genes in nonlesional rebiopsies (NonLes_rebiopsy compared with Ctrl) against FC of genes in upper buttock wound healing (UB_Week6 compared with UB_Ctrl). All genes are represented in gray. Wound healing signature (SWH) genes are colored red, and KAS genes are colored blue. Genes in green are wound-healing–specific genes not significant in any of the keloid groups. KAS genes with the largest fold changes in either direction are labeled by name. Ctrl, control; FC, fold change; KAS, keloid-associated signature; SWH, shared wound healing; UB, upper buttock.
KAS gene COL2A1 as a potential biomarker of mature and developing keloids
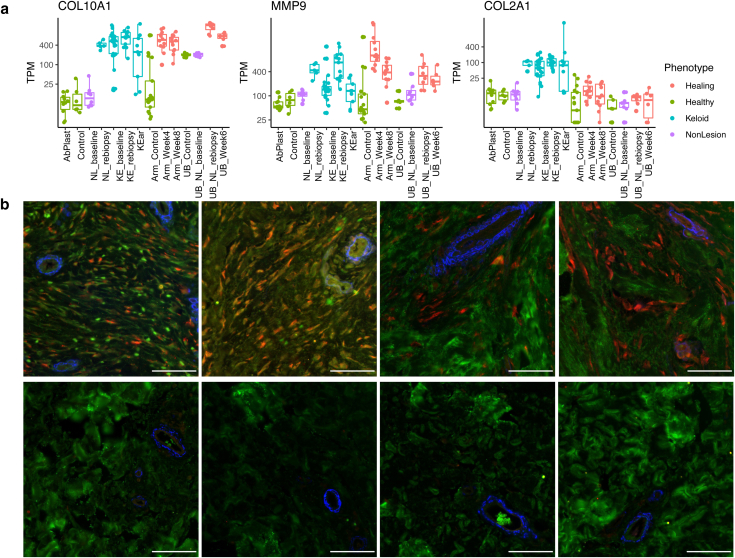
The 12 most prominent KAS genes include KCNC1, a potassium voltage-gated channel gene; NPTX2, a neural protein; ITGA8, an integrin protein; TMEM132E, a transmembrane protein; and PDPR, a regulatory subunit of mitochondrial pyruvate dehydrogenase phosphatase. Carrying out comprehensive molecular validation for all KAS genes is outside the scope of this paper. We placed our focus on COL2A1, a gene coding for the pro-alpha1(II) chain that makes up the mature triple-stranded type II collagen. COL2A1 is not a gene that is usually expressed in the skin. Type II collagen is found primarily in cartilage, bone, the vitreous humor, and the nucleus pulposus. We confirmed that COL2A1 was only upregulated in keloid groups, contrasting with classic wound healing genes such as COL10A1 and matrix metalloproteinase 9 gene MMP9 (Figure 11a). To further validate this finding, four healthy skin and keloid skin samples were stained for COL2A1, showing clear COL2A1 expression in keloid skin that was consistently absent in healthy skin (Figure 11b).
Figure 11.
Immunohistochemistry of COL2A1 in healthy skin and ear keloids. (a) Boxplots of TPM values of COL2A1, COL10A1, and MMP9 across comparison groups, colored by tissue type. The y-axis is in log scale. (b) Formalin-fixed, paraffin-embedded specimens from keloid (top, n = 4) or abdominoplasty (bottom, n = 4) were subject to immunohistochemical colocalization of Col2 (red), Col1 (green), and α-SMA (blue). Col2 and subcellular Col1 were found exclusively in keloid, although they were associated with different cells. Bar = 50 μm. α-SMA, α-smooth muscle actin; Col1, collagen type 1; Col2, collagen type 2; MMP9, matrix metalloproteinase 9; TPM, transcripts per million.
Discussion
COL2A1: potential role in keloid development and maintenance
COL2A1 is a component of type II collagen, an important molecule in bone formation, normal joint function, and the development of the eye and ear (Gregersen and Savarirayan, 1993). It is involved in collagen chain trimerization, endochondral ossification, and signaling by PDGF. Alterations in COL2A1 have been linked to bone and cartilage diseases such as chondrosarcoma (Tarpey et al., 2013), spondyloemetaphyseal dysplasia (Matsubayashi et al., 2013), and kniest dysplasia (Spranger et al., 1994).
The increased expression and accumulation of COL2A1 in keloid tissue may be due to the disordered differentiation of cell types in lesional skin. Report of elevation of chondrocyte/osteoblast marker genes, including SOX9 and CBFA1, in keloids has been previously described (Naitoh et al., 2005). Upregulation of genes involved in bone and cartilage formation has also been published (Inui et al., 2011) in African American subjects (BMP1, RUNX2, and CDH11) (Fuentes-Duculan et al., 2017) and more recently in another cohort (Lin et al., 2022). It must be noted that Lin et al. (2022) specifically reported no change in COL2A1 levels between control and keloid dermal tissues, with extremely low expression in all samples. Although the patient population source in the study by Lin et al. (2022) is different, whether these differences caused the discrepancy in COL2A1 levels requires further investigation. There may be some support for COL2A1 expression in keloid tissue. A proteomic study identified cartilage-like protein composition of keloid scar extracellular matrix and included COL2A1 as one of a few keloid-unique proteins. The authors suggested in their paper that fibrillar collagen network is different in keloids (Barallobre-Barreiro et al., 2019). Therefore, one hypothesis for the involvement of COL2A1 in keloid development may be the incorporation of COL2A1 into collagen trimers (incorporation of COL2A1 + COL1A1), leading to greater stability and stiffness of fibrillar collagens, perhaps owing to higher cross-linking; negatively impacting the turnover rate; and leading to increased deposition over time.
COL2A1 was also suggested as a biomarker of melanoma tumor–repopulating cells (Talluri et al., 2020), and the tumor-like nature of keloids is intriguing considering this shared gene with known skin cancer. Because COL2A1 is not commonly expressed in healthy skin, it has the potential to serve as a biomarker for keloids, but substantial further work will be required to validate this result.
Wound-healing stage in keloid signature
Gene set enrichment analysis of the mature keloid signature agreed with trends previously reported (Diaz et al., 2020; Fuentes-Duculan et al., 2017; Wu et al., 2020) and support the current understanding of keloid histopathology (Limandjaja et al., 2020). The classic model of healthy wound healing is divided into four overlapping phases: hemostasis, inflammatory, proliferative, and maturation (Landén et al., 2016; Nurden et al., 2008), generally lasting less than a month in total, although it can be up to a year depending on the size of the wound and the condition of the patient. In contrast, keloid development takes 6 weeks to 12 months to be noticeable, and wound sites can appear visually healed before keloid growth. This study includes skin biopsies on day 0 and rebiopsies 47–107 days later, representing tissue that should be well into the maturation phase where the skin is healing correctly. Although the classic model of wound healing describes the process as discreet and stepwise, there is no apparent dominant phase of healing. Inflammation, proliferation, and matrix remodeling pathways were all enriched in the keloid signature as well as in healing skin. The large overlap between keloids and wound-healing signatures 4–8 weeks after injury in healthy skin supports the hypothesis that keloids are, at least in part, persistent dysregulations of the general wound-healing process.
Cell types in keloid development
Keloid growth is the result of the dysregulation of multiple cell types in healing skin. Most notably, fibroblasts play a key role in collagen secretion and fibrosis and contribute significantly to keloid growth (Ashcroft et al., 2013; Hsu et al., 2018). Efforts to target and suppress fibroblasts in keloids have led to strategies such as regulating fibroblasts for scarless wound healing (Zou et al., 2021) or inhibiting keloid-derived fibroblast growth and migration with LY2109761 (Wang et al., 2021). In tandem, keratinocytes secrete factors that modulate fibroblast collagen production (Alghamdi et al., 2020), and mast cells play an important role in dermal scarring (Ud-Din et al., 2020) by interacting with fibroblasts in epithelial growth and regeneration (Artuc et al., 2002). Bulk deconvolution showed unsurprising increases in fibroblasts, endothelial cells, and M2 macrophages/dendritic cells in keloid skin. The inferred decrease in keratinocytes was unexpected because morphologically, the epidermal layer is thickened in comparison with that of healthy skin. This could be due to differences in keratinocyte activity or RNA-extraction efficiency between healthy and lesional skin. Because bulk sequencing reports relative abundance in whole tissue, analyzing skin layers separately may also clarify this observation. The deconvolution method is limited by the cell types present in the reference single-cell data and relies on the availability of robust cell type–specific gene markers. In addition, proportional changes are not indicative of absolute cell count changes, and the lower detection limit of deconvolution places large error margins on rarer cell types below 5% abundance. Finally, the varied skin locations and ancestries present in this multistudy comparison may account for some of the patterns observed in addition to true differences between developing/mature keloids and healthy healing skin. For example, mast cells are enriched in keloid groups compared with those in the control but not in comparison with those in AbPlast. They are also elevated in upper buttock wound healing but not in arm wound healing. There may be inherent differences in mast cell levels in the skin from different parts of the body, or it could be linked to a propensity to develop keloids. With additional work and future datasets, these observations will be further elucidated. Efforts to target responsible cell types, combinations thereof, or the cell–cell communication signaling molecules may prove fruitful as ongoing areas of research.
Age and genetics in heterogeneity in mature keloids
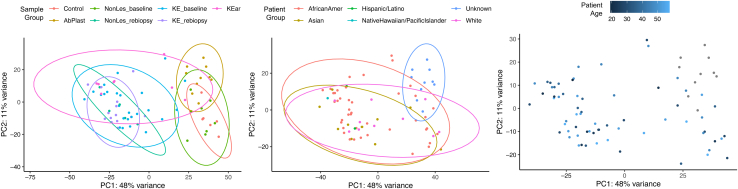
Keloid development is age linked. There is likely a role for hormonal levels being associated with low keloid growth in prepubescent individuals (Chike-Obi et al., 2009; Ibrahim et al., 2020), whereas slower wound healing and regeneration may be associated with reduced keloid growth in mature skin (Ashcroft et al., 1998; Gerstein et al., 1993; Gunin et al., 2011; Solé-Boldo et al., 2020). Our study only included adults, the majority of whom fall between ages of 20 and 50 years. Analyzing samples by age did not observe the clustering of keloid samples (Figure 12).
Figure 12.
PCAs colored by biopsy group, race, and age. PC, principal component; PCA, principal component analysis.
Studies suggested that familial tendency to develop keloids may be inherited in an autosomal dominant pattern with incomplete penetrance (Marneros et al., 2001; Yan et al., 2007), and GWASs have shown that multiple loci are correlated with a genetic susceptibility for keloid development (Chen et al., 2007, 2006; Marneros et al., 2004; Nakashima et al., 2010; Yan et al., 2007; Zhu et al., 2013). However, these investigations do not sufficiently explain the contribution of ancestry to keloid biology (Chen et al., 2006; Chike-Obi et al., 2009; Clark et al., 2009; Huang et al., 2020; Russell et al., 2010). In our study, there was insufficient power to carry out differential analyses controlling for ancestry. However, we also did not observe the clustering of keloid samples by self-identified race or ethnicity (Figure 12).
Summary
The pathogenesis of keloids and the means of effective treatment and prevention have remained elusive. This study provides the largest keloid bulk-RNA sequencing dataset published to date, with baseline and injured lesional and nonlesional rebiopsies and subjects spanning diverse racial and ethnic groups. Rebiopsies of wounded nonlesional and lesional skin reveal a developing keloid signature in nonlesional skin after trauma, nearly identical to that of the mature keloid. Comparison of the mature and developing keloid signatures with those of healthy wound healing allows the definition of a robust SWH signature and a keloid-associated signature. Few biomarkers have been reported for keloids, with published genes limited to CD138 (Bagabir et al., 2016), matrix metalloproteinase 9 gene MMP19, and CGRP (Suarez et al., 2015). COL2A1 is, to our knowledge, a previously unreported addition to this short list, but a cross-comparison of the complete SWH and KAS lists with future studies, in conjunction with molecular validation, may identify the potential mechanisms and provide clarity to identify best targets for clinical treatment.
Materials and Methods
Participants
The study was approved by the Northwestern University Institutional Review Board (STU00203462) and registered on Clinicaltrials.gov (NCT03228693). A total of 48 subjects were recruited from 2017 to 2020 with written, informed consent, with a total of 31 finally analyzed. Inclusion criteria included adults aged >18 years with keloid scar that was untreated or who were treatment free for a minimum of 6 months before enrollment. Keloid diagnosis was confirmed by a board-certified dermatologist. Participants were compensated for their time and participation in the study. All study visits were conducted at the Northwestern University Feinberg School of Medicine Department of Dermatology (Chicago, IL). The Northwestern University Dermatology Tissue and Acquisition and Repository project (institutional review board number STU00009443), part of the Skin Biology and Diseases Resource-Based Center, was used to collect an additional 6 ear keloid tissue samples and 10 abdominoplasty tissue samples.
Keloid lesional and nonlesional skin sample collection
Photographs and measurements of keloid (lesional) scars were taken before biopsy. The site of lesional biopsy was restricted to the trunk or arms. A total of 6-mm skin punch biopsies to subcutaneous fat were performed using standard clinical practice. Biopsies were taken at the lesion periphery (keloid edge) fully inclusive of the keloid scar (no adjacent normal skin). The biopsy was immediately bisected and placed into RNALater solution and stored at 4 °C for 24–48 hours and then transferred to –80 °C for storage before processing.
Earlobe keloid excision procedure and handling
Earlobe keloid excision was performed using a standard technique. The excised tissue was trisected. One 6-mm punch was placed in RNALater for RNA sequencing, the second piece was frozen in liquid nitrogen for proteomics, and the third piece was embedded in optimal cutting temperature media and frozen on dry ice for histology. All pieces were stored at –80 °C before processing.
Immunohistochemistry on skin paraffin sections
Specimens for histology were fixed in 10% neutral buffered formalin for 36 hours, processed to paraffin blocks, and cut (5 μm) onto glass slides for immunohistochemical processing on a Leica Bond Rx Stainer. In brief, slides were pretreated for epitope retrieval by incubating them in a citrate-based buffer (pH 6) for 20 minutes at 100 °C; slides were then treated with a 30-minute protein blocking step before incubation with the primary antibody cocktail for 1 hour, followed by a 1-hour incubation with the secondary antibody cocktail. After staining was completed, slides were coverslipped (1.5 mm glass) using an aqueous mounting media with DAPI and then imaged on a Zeiss Axioscan. Images were qualitatively assessed for differential COL2A1 expression. Reagent and antibody specifications are listed in Table 2.
Table 2.
Table of Antibodies
| Category | Target | Host | Isotype [Clone] | Vendor | Catalog Number | Dilution | Conjugate |
|---|---|---|---|---|---|---|---|
| 1o Antibody | Col1 | Mouse | IgG1 [COL-1] | Abcam | AB6308 | 1:200 | |
| 1o Antibody | Col2 | Rabbit | Polyclonal | Invitrogen | PA5-85108 | 1:100 | |
| 1o Antibody | α-SMA | Mouse | IgG2a [1A4] | R&D Systems | IC1420S | 1:75 | Alexa 750 |
| 2o Antibody | Mouse IgG1 | Goat | Jackson Immunoresearch | 115-165-205 | 1:333 | Cy3 | |
| 2o Antibody | Mouse IgG2a | Goat | Jackson Immunoresearch | 115-605-206 | 1:333 | Alexa 647 |
| Category | Description | Vendor | Catalog Number | Notes |
|---|---|---|---|---|
| Ancillary | PowerVision IHC/ISH Super Blocking | Leica Biosystems | BB02-110 | Protein Block |
| Ancillary | Antibody Diluent | Leica Biosystems | AR9352 | |
| Ancillary | Fluoro-Gel II with DAPI | Electron Microscopy Sciences | 17985-51 | Mounting Media |
| Consumable | Glass coverslips, 25 × 60 × 1.5 mm | Leica Biosystems | 3800161 |
Bulk RNA sequencing
Strand-specific RNA-sequencing libraries were prepared from 500 ng RNA using KAPA stranded mRNA-Seq Kit (Kapa Biosystems, Wilmington, MA). Twelve-cycle PCR was performed to amplify libraries. Sequencing was performed on Illumina HiSeq2500 (Illumina, San Diego, CA) by multiplexed paired-read run with 33 cycles. Raw sequence data (BCL files) were converted to FASTQ format through Illumina Casava 1.8.2. Reads were decoded on the basis of their barcodes, and read quality was evaluated with FastQC. Reads were mapped to the human genome (National Center for Biotechnology Information B37.3) and the University of California Santa Cruz gene model using ArrayStudio software (OmicSoft, Cary, NC) allowing two mismatches. Reads mapped to the sense-strand exons of a gene were summed at the gene level.
Publicly available wound healing datasets
GSE152781 contained volunteer skin taken from the inner arm at baseline (Arm_Control, n = 17) and injured skin taken from the same location 4 weeks later (Arm_Week4, n = 12) and again 8 weeks later (Arm_Week8, n = 12). GSE113619 contained skin from the upper buttock. Samples were collected from volunteers without keloid history at baseline (UB_Control, n = 5) and injured skin 6 weeks later (UB_Week6, n = 6) as well as nonlesional skin from subjects with a history of keloids at baseline (UB_NonLes_baseline) and 6 weeks later (UB_NonLes_rebiopsy).
Bulk RNA-sequencing differential analysis
Differential gene expression analysis was carried out with DESeq2, version 1.30.0. Un-normalized, rounded counts at the gene level as summarized by ArrayStudio software were used as input. Genes XIST and KDM5D were used as female- and male-specific markers, respectively, to assign sex where necessary to include as a covariate in the analysis model. Genes were prefiltered for a minimum of 10 reads in 75% of the sample in any one of the comparison groups. Size factors were estimated per sample using the median-of-ratios method to account for intersample sequencing depth variations. Hypothesis testing was carried out using the Wald test, with multiple testing correction by Benjamini–Hochberg method. Significance was defined as adjusted P < 0.05 and absolute fold change > 1.5.
Gene set enrichment analysis
All genes were sorted using the signal-to-noise ratio output from DESeq2 analysis. The presorted gene lists for each comparison were used as input into gene set enrichment analysis using the fgsea R package. Gene sets (Hallmark h.all, version 7.1, and Reactome c2.cp.reactome, version 7.1) were obtained from the MSigDB (Molecular Signature Database) (Subramanian et al., 2005).
Bulk deconvolution
For AdRoit 2.0 (Yang et al., 2021), reannotated and reanalyzed public Seurat object was downloaded from GSE163973. Seurat’s FindAllMarkers function was used to generate cell-type markers. Cell types were reannotated for deconvolution reference generation (1,000 genes). For CIBERSORTx (Newman et al., 2019), bulk RNA sequencing was conducted through authorized access from https://cibersortx.stanford.edu/. Briefly, a signature matrix was created from the reannotated and reanalyzed public single-cell RNA-sequencing data GSE163973. The mixture file was created using the raw counts file from the bulk RNA sequencing without quantile normalization. A total of 100 permutations were selected to run the algorithm. For MuSiC2 (Wang et al., 2019b), deconvolution was carried out according to the publicly available vignette on the GitHub page.
Data availability statement
Data access
Gene count table and accompanying metadata table from bulk-RNA sequencing are provided in Supplementary Data. Fastq files are publicly available on https://www.ncbi.nlm.nih.gov bioproject PRJNA905476.
Code availability
No new algorithms were developed for this manuscript. Scripts generated for analysis and figures are available upon request.
ORCIDs
Yuan O. Zhu: http://orcid.org/0000-0001-9419-6291
Scott MacDonnell: http://orcid.org/0000-0001-8380-1572
Theodore Kaplan: http://orcid.org/0000-0001-5825-903X
Chien Liu: http://orcid.org/0000-0003-1674-2001
Yasmeen Ali: http://orcid.org/0000-0002-8055-7815
Stephanie M. Rangel: http://orcid.org/0000-0002-1206-9561
Madeleine Belback: http://orcid.org/0000-0003-2059-0864
Matthew F. Wipperman: http://orcid.org/0000-0003-1436-3366
Daphne S. Sun: http://orcid.org/0000-0002-7647-7169
Ziyou Ren: http://orcid.org/0000-0001-7127-4197
Xiaolong Alan Zhou: http://orcid.org/0000-0002-6177-2472
Gabor Halasz: http://orcid.org/0000-0002-3332-033X
Lori Morton: http://orcid.org/0000-0002-5214-5784
Roopal V. Kundu: http://orcid.org/0000-0002-7509-3606
Conflict of Interest
YOZ, SM, TK, JL, MFW, DSS, GH, and LM are employees and stockholders of Regeneron Pharmaceuticals. RVK is a principal investigator for miRagen Therapeutics, Pfizer, Regeneron Pharmaceuticals, Avita Medical Americas, and Incyte Corporation and receives grant support and funding to conduct research as a full-time academic faculty member. The remaining authors state no conflict of interest.
Acknowledgments
The authors would like to acknowledge the study participants who generously enabled this research work. Research reported in this publication was supported, in part, by the National Institutes of Health's National Center for Advancing Translational Sciences (grant number UL1TR001422) (Northwestern). Research reported in this publication was supported by Northwestern University Skin Biology & Diseases Resource-Based Center of the National Institutes of Health under award number P30AR075049. This work was supported by the Northwestern University NUSeq Core Facility. We gratefully acknowledge Regeneron’s Molecular Profiling Labs for RNA sequencing of all samples, and DNA Core for sample preparation.
Author Contributions
Conceptualization: RVK, SM, GH, LM; Data Curation: SM, SMR, MB, YA; Formal Analysis: YOZ, GH, SM, ZR, DSS; Funding Acquisition: SM, LM, RVK; Investigation: TK, CL, YOZ, GH, MFW, SMR, MB, XAZ, YA, RVK; Methodology: TK, CL, YOZ, GH, DSS, ZR, RVK; Project Administration: SM, SMR; Resources: SM, TK, CL, YA, SMR, YOZ, GH; Software: YOZ, MFW, GH, ZR, DSS; Supervision: SM, SMR, LM, RVK; Validation: SM, TK, CL, YOZ, GH, DSS, ZR, LM, RVK; Visualization: SM, YOZ, GH, DSS, LM, RVK; Writing - Original Draft Preparation: YOZ, SM, GH, RVK; Writing - Review and Editing: YOZ, SM, TK, CL, MFW, SMR, MB, DSS, XAZ, YA, RVK
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2023.100211
Supplementary material is linked to the online version of the paper at www.jidonline.org and at https://doi.org/10.1016/j.jid.2023.100211.
Supplementary Materials
References
- Alghamdi M.A., Al-Eitan L.N., Stevenson A., Chaudhari N., Hortin N., Wallace H.J., et al. Secreted factors from keloid keratinocytes modulate collagen deposition by fibroblasts from normal and fibrotic tissue: a pilot study. Biomedicines. 2020;8:200. doi: 10.3390/biomedicines8070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artuc M., Steckelings U.M., Henz B.M. Mast cell-fibroblast interactions: human mast cells as source and inducers of fibroblast and epithelial growth factors. J Invest Dermatol. 2002;118:391–395. doi: 10.1046/j.0022-202x.2001.01705.x. [DOI] [PubMed] [Google Scholar]
- Ashcroft G.S., Horan M.A., Ferguson M.W. Aging alters the inflammatory and endothelial cell adhesion molecule profiles during human cutaneous wound healing. Lab Invest. 1998;78:47–58. [PubMed] [Google Scholar]
- Ashcroft K.J., Syed F., Bayat A. Site-specific keloid fibroblasts alter the behaviour of normal skin and normal scar fibroblasts through paracrine signalling. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagabir R.A., Syed F., Shenjere P., Paus R., Bayat A. Identification of a potential molecular diagnostic biomarker in keloid disease: Syndecan-1 (CD138) is overexpressed in keloid scar tissue. J Invest Dermatol. 2016;136:2319–2323. doi: 10.1016/j.jid.2016.06.623. [DOI] [PubMed] [Google Scholar]
- Barallobre-Barreiro J., Woods E., Bell R.E., Easton J.A., Hobbs C., Eager M., et al. Cartilage-like composition of keloid scar extracellular matrix suggests fibroblast mis-differentiation in disease. Matrix Biol Plus. 2019;4 doi: 10.1016/j.mbplus.2019.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet U., Blalock T.W. Keloids: a review of etiology, prevention, and treatment. J Clin Aesthet Dermatol. 2020;13:33–43. [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gao J.H., Liu X.J., Yan X., Song M. Characteristics of occurrence for Han Chinese familial keloids. Burns. 2006;32:1052–1059. doi: 10.1016/j.burns.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chen Y., Gao J.H., Yan X., Song M., Liu X.J. [Location of predisposing gene for one Han Chinese keloid pedigree] Zhonghua Zheng Xing Wai Ke Za Zhi. 2007;23:137–140. [PubMed] [Google Scholar]
- Chike-Obi C.J., Cole P.D., Brissett A.E. Keloids: pathogenesis, clinical features, and management. Semin Plast Surg. 2009;23:178–184. doi: 10.1055/s-0029-1224797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.A., Turner M.L., Howard L., Stanescu H., Kleta R., Kopp J.B. Description of familial keloids in five pedigrees: evidence for autosomal dominant inheritance and phenotypic heterogeneity. BMC Dermatol. 2009;9:8. doi: 10.1186/1471-5945-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C.C., Hu Y.F., Zhu D.H., Cheng Q., Gu J.J., Feng Q.L., et al. Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat Commun. 2021;12:3709. doi: 10.1038/s41467-021-24110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Tan K., He H., Xu H., Cueto I., Pavel A.B., et al. Keloid lesions show increased IL-4/IL-13 signaling and respond to Th2-targeting dupilumab therapy. J Eur Acad Dermatol Venereol. 2020;34:e161–e164. doi: 10.1111/jdv.16097. [DOI] [PubMed] [Google Scholar]
- Fuentes-Duculan J., Bonifacio K.M., Suárez-Fariñas M., Kunjravia N., Garcet S., Cruz T., et al. Aberrant connective tissue differentiation towards cartilage and bone underlies human keloids in African Americans. Exp Dermatol. 2017;26:721–727. doi: 10.1111/exd.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauglitz G.G., Korting H.C., Pavicic T., Ruzicka T., Jeschke M.G. Hypertrophic scarring and keloids: pathomechanisms and current and emerging treatment strategies. Mol Med. 2011;17:113–125. doi: 10.2119/molmed.2009.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A.D., Phillips T.J., Rogers G.S., Gilchrest B.A. Wound healing and aging. Dermatol Clin. 1993;11:749–757. [PubMed] [Google Scholar]
- Gregersen PA, Savarirayan R, Adam MP, Mirzaa GM, Pagon RA, Wallace SE, et al. Type II collagen disorders overview. In: GeneReviews. Seattle, WA: University of Washington Press; 1993. [PubMed]
- Gunin A.G., Kornilova N.K., Vasilieva O.V., Petrov V.V. Age-related changes in proliferation, the numbers of mast cells, eosinophils, and CD45-positive cells in human dermis. J Gerontol A Biol Sci Med Sci. 2011;66:385–392. doi: 10.1093/gerona/glq205. [DOI] [PubMed] [Google Scholar]
- Hsu C.K., Lin H.H., Harn H.I., Ogawa R., Wang Y.K., Ho Y.T., et al. Caveolin-1 controls hyperresponsiveness to mechanical stimuli and fibrogenesis-associated RUNX2 activation in keloid fibroblasts. J Invest Dermatol. 2018;138:208–218. doi: 10.1016/j.jid.2017.05.041. [DOI] [PubMed] [Google Scholar]
- Huang C., Wu Z., Du Y., Ogawa R. In: Textbook on scar management: state of the art management and emerging technologies. Téot L., Mustoe T.A., Middelkoop E., Gauglitz G.G., editors. Springer International Publishing; Cham, Switzerland: 2020. The epidemiology of keloids; pp. 29–35. [PubMed] [Google Scholar]
- Ibrahim N.E., Shaharan S., Dheansa B. Adverse effects of pregnancy on keloids and hypertrophic scars. Cureus. 2020;12 doi: 10.7759/cureus.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui S., Shono F., Nakajima T., Hosokawa K., Itami S. Identification and characterization of cartilage oligomeric matrix protein as a novel pathogenic factor in keloids. Am J Pathol. 2011;179:1951–1960. doi: 10.1016/j.ajpath.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landén N.X., Li D., Ståhle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell Mol Life Sci. 2016;73:3861–3885. doi: 10.1007/s00018-016-2268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limandjaja G.C., Niessen F.B., Scheper R.J., Gibbs S. The keloid disorder: heterogeneity, histopathology, mechanisms and models. Front Cell Dev Biol. 2020;8:360. doi: 10.3389/fcell.2020.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Zhang G., Peng R., Zhao M., Li H. Increased expression of bone/cartilage-associated genes and core transcription factors in keloids by RNA sequencing. Exp Dermatol. 2022;31:1586–1596. doi: 10.1111/exd.14630. [DOI] [PubMed] [Google Scholar]
- Liu X., Chen W., Zeng Q., Ma B., Li Z., Meng T., et al. Single-cell RNA-sequencing reveals lineage-specific regulatory changes of fibroblasts and vascular endothelial cells in keloids. J Invest Dermatol. 2022;142:124–135.e11. doi: 10.1016/j.jid.2021.06.010. [DOI] [PubMed] [Google Scholar]
- Mari W., Alsabri S.G., Tabal N., Younes S., Sherif A., Simman R. Novel insights on understanding of keloid scar: article review. J Am Coll Clin Wound Spec. 2015;7:1–7. doi: 10.1016/j.jccw.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marneros A.G., Norris J.E., Olsen B.R., Reichenberger E. Clinical genetics of familial keloids. Arch Dermatol. 2001;137:1429–1434. doi: 10.1001/archderm.137.11.1429. [DOI] [PubMed] [Google Scholar]
- Marneros A.G., Norris J.E., Watanabe S., Reichenberger E., Olsen B.R. Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. J Invest Dermatol. 2004;122:1126–1132. doi: 10.1111/j.0022-202X.2004.22327.x. [DOI] [PubMed] [Google Scholar]
- Matsubayashi S., Ikema M., Ninomiya Y., Yamaguchi K., Ikegawa S., Nishimura G. COL2A1 mutation in spondylometaphyseal dysplasia Algerian type. Mol Syndromol. 2013;4:148–151. doi: 10.1159/000346644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N.M., Aoki M., Okubo Y., Kuwahara K., Eura S., Dohi T., et al. Gene expression profile of isolated dermal vascular endothelial cells in keloids. Front Cell Dev Biol. 2020;8:658. doi: 10.3389/fcell.2020.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naitoh M., Kubota H., Ikeda M., Tanaka T., Shirane H., Suzuki S., et al. Gene expression in human keloids is altered from dermal to chondrocytic and osteogenic lineage. Genes Cells. 2005;10:1081–1091. doi: 10.1111/j.1365-2443.2005.00902.x. [DOI] [PubMed] [Google Scholar]
- Nakashima M., Chung S., Takahashi A., Kamatani N., Kawaguchi T., Tsunoda T., et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–771. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurden A.T., Nurden P., Sanchez M., Andia I., Anitua E. Platelets and wound healing. Front Biosci. 2008;13:3532–3548. doi: 10.2741/2947. [DOI] [PubMed] [Google Scholar]
- Onoufriadis A., Hsu C.K., Ainali C., Ung C.Y., Rashidghamat E., Yang H.S., et al. Time series integrative analysis of RNA sequencing and microRNA expression data reveals key biologic wound healing pathways in keloid-prone individuals. J Invest Dermatol. 2018;138:2690–2693. doi: 10.1016/j.jid.2018.05.017. [DOI] [PubMed] [Google Scholar]
- Russell S.B., Russell J.D., Trupin K.M., Gayden A.E., Opalenik S.R., Nanney L.B., et al. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–2496. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé-Boldo L., Raddatz G., Schütz S., Mallm J.P., Rippe K., Lonsdorf A.S., et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol. 2020;3:188. doi: 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger J., Menger H., Mundlos S., Winterpacht A., Zabel B. Kniest dysplasia is caused by dominant collagen II (COL2A1) mutations: parental somatic mosaicism manifesting as Stickler phenotype and mild spondyloepiphyseal dysplasia. Pediatr Radiol. 1994;24:431–435. doi: 10.1007/BF02011911. [DOI] [PubMed] [Google Scholar]
- Suarez E., Syed F., Alonso-Rasgado T., Bayat A. Identification of biomarkers involved in differential profiling of hypertrophic and keloid scars versus normal skin. Arch Dermatol Res. 2015;307:115–133. doi: 10.1007/s00403-014-1512-4. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talluri B., Amar K., Saul M., Shireen T., Konjufca V., Ma J., et al. COL2A1 is a novel biomarker of melanoma tumor repopulating cells. Biomedicines. 2020;8:360. doi: 10.3390/biomedicines8090360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Khumalo N., Bayat A. Understanding keloid pathobiology from a quasi-neoplastic perspective: less of a scar and more of a chronic inflammatory disease with cancer-like tendencies. Front Immunol. 2019;10:1810. doi: 10.3389/fimmu.2019.01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey P.S., Behjati S., Cooke S.L., Van Loo P., Wedge D.C., Pillay N., et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat Genet. 2013;45:923–926. doi: 10.1038/ng.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Din S., Wilgus T.A., Bayat A. Mast cells in skin scarring: a review of animal and human research. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.552205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Din S., Wilgus T.A., McGeorge D.D., Bayat A. Pre-emptive priming of human skin improves cutaneous scarring and is superior to immediate and delayed topical anti-scarring treatment post-wounding: a double-blind randomised placebo-controlled clinical trial. Pharmaceutics. 2021;13:510. doi: 10.3390/pharmaceutics13040510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wu H., Xiao Z., Dong X. Expression profiles of lncRNAs and circRNAs in keloid. Plast Reconstr Surg Glob Open. 2019;7:e2265. doi: 10.1097/GOX.0000000000002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gu C., Shang F., Jin R., Zhou J., Gao Z. Inhibitory effect of the LY2109761 on the development of human keloid fibroblasts. Anal Cell Pathol (Amst) 2021;2021 doi: 10.1155/2021/8883427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Del Duca E., Espino M., Gontzes A., Cueto I., Zhang N., et al. RNA sequencing keloid transcriptome associates keloids with Th2, Th1, Th17/Th22, and JAK3-skewing. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.597741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Gu S., Huang X., Ren J., Gu Y., Wei C., et al. The role of macrophages in the formation of hypertrophic scars and keloids. Burns Trauma. 2020;8 doi: 10.1093/burnst/tkaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Gao J.H., Chen Y., Song M., Liu X.J. [Preliminary linkage analysis and mapping of keloid susceptibility locus in a Chinese pedigree] Zhonghua Zheng Xing Wai Ke Za Zhi. 2007;23:32–35. [PubMed] [Google Scholar]
- Yang T., Alessandri-Haber N., Fury W., Schaner M., Breese R., LaCroix-Fralish M., et al. AdRoit is an accurate and robust method to infer complex transcriptome composition. Commun Biol. 2021;4:1218. doi: 10.1038/s42003-021-02739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Wu B., Li P., Wang J., Tang H., Liu Y., et al. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M.L., Teng Y.Y., Wu J.J., Liu S.Y., Tang X.Y., Jia Y., et al. Fibroblasts: heterogeneous cells with potential in regenerative therapy for scarless wound healing. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.713605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data access
Gene count table and accompanying metadata table from bulk-RNA sequencing are provided in Supplementary Data. Fastq files are publicly available on https://www.ncbi.nlm.nih.gov bioproject PRJNA905476.
Code availability
No new algorithms were developed for this manuscript. Scripts generated for analysis and figures are available upon request.