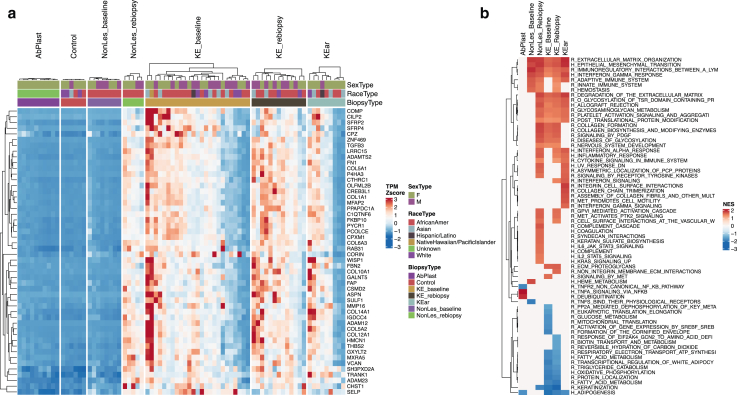
Figure 2.
Keloid signature and pathway enrichment analysis. (a) Data showing the top 50 genes in keloid signature by P-value (KE_b compared with Ctrl). Heatmap of TPM Z scores (percentile capped at 5th and 95th) show that this signature is reflected across keloid groups (KE_rebiopsy and KEar) as well as post-trauma nonlesional skin (NonLes_rebiopsy) but is absent in both healthy skin groups (AbPlast and Ctrl) and baseline nonlesional skin (NonLes_baseline). Some heterogeneity of signature intensity is observed within each keloid group. Biological sex, race, and biopsy type are indicated in the figure column header. (b) Select GSEA results, ordered by maximum absolute NES across comparison groups. Missing values indicate nonsignificance. Pathways tested came from Hallmark (denoted as H_) or Reactome (denoted as R_) databases. Repetitive and irrelevant pathways were excluded. Ctrl, control; GSEA, gene set enrichment analysis; NES, normalized enrichment score; TPM, transcripts per million.