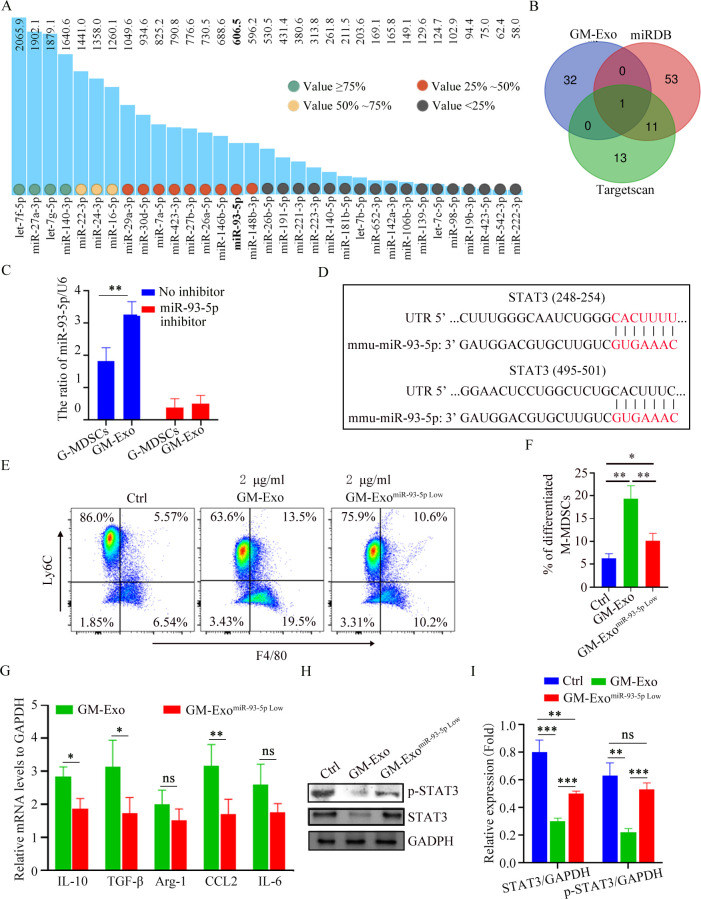
Figure 5.
GM-Exo downregulate the STAT3 activity in M-MDSCs via miR-93–5 p. (A) MiRNAs with expression value are greater than 50 in GM-Exo. (B) Venn diagram of predicted miRNA by Targetscan and miRDB databases. (C) The relative expression of miR-93–5 p was analyzed using qRT-PCR (n=3). (D) Binding sites between the STAT3 3’ UTR and miR-93–5 p were identified by Targetscan. (E) M-MDSCs were cultured for 72 hours in the presence of Sup and GM-Exo. The percentage of F4/80+Ly6Clow cell were detected by FACS (n=3). (F) Summary graphs of the percentage of differentiated M-MDSCs to total M-MDSCs (n=3). (G) mRNA expression of cytokines in M-MDSCs was analyzed by qRT-PCR (n=3). (H) IB analysis of p-STAT3 and STAT3 levels in exosomes-treated M-MDSC for 48 hours. (I) Summary graphs of relative expression of p-STAT3 and STAT3 in exosomes-treated M-MDSCs (n=3). Statistical analyses were performed using unpaired t-tests. M-MDSCs, monocytic myeloid-derived suppressor cells. (*p<0.05, **p<0.01, ***p<0.001, ns, not significant, one-way ANOVA test or unpaired t test; error bars, SD).