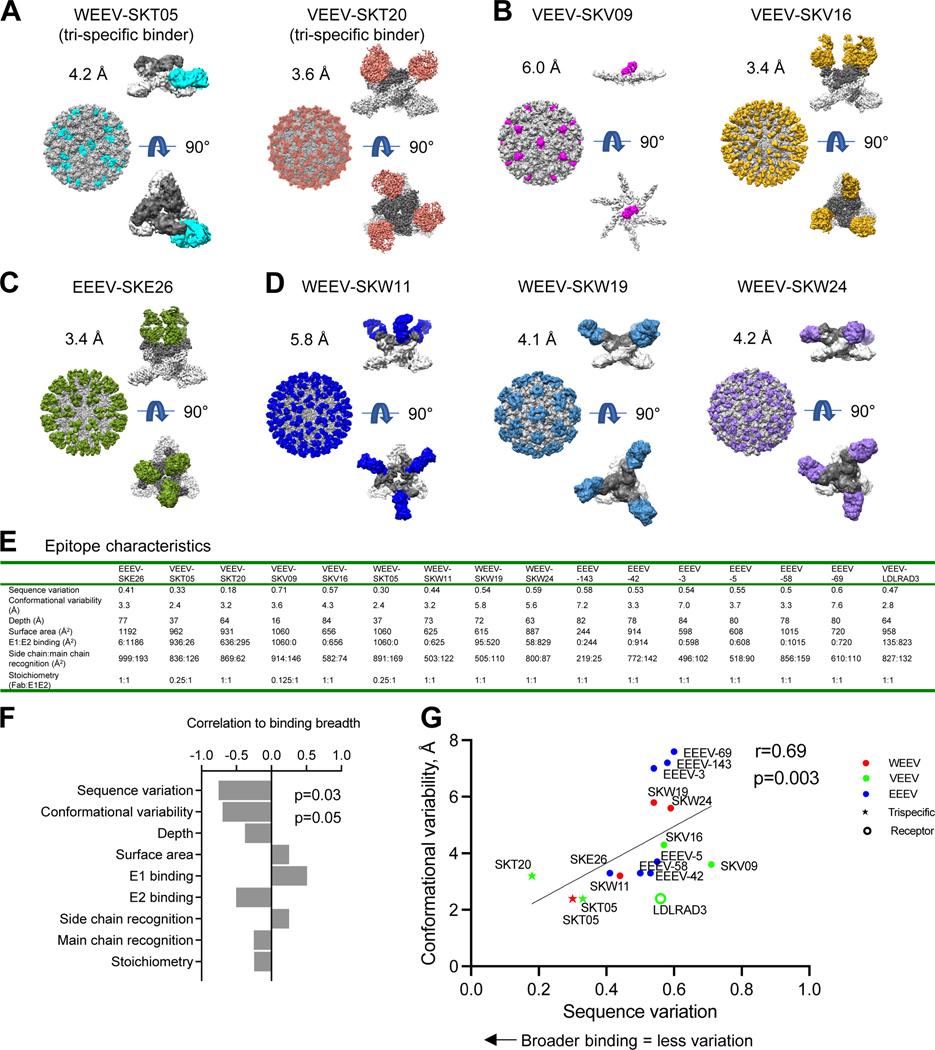
Figure 4. Cryo-EM structures of neutralizing antibodies with VLPs reveal that sequence variation and conformation variability inversely correlate with broad recognition.
(A-D) Cryo-EM structures of select (A) triple-specific antibodies, (B) α-VEEV antibodies, (C) α-EEEV antibodies, or (D) α-WEEV antibodies. Each antibody neutralized at least one pseudovirus. For each antibody-VLP complex, the entire complex is shown with VLP, with VLP in gray and Fabs colored. The E1E2 spike with bound Fab is also shown E1 in light gray, E2 in dark gray, and Fab colored; a side view is shown at top, and under it, a 90° rotation viewing down the spike molecular 3-fold axis (note that for VEEV-SKV09, this Fab binds directly to E1 at the 2-fold axis of the VLP, and this interaction is shown from side and along 2-fold axis). (E) Epitope characteristics are calculated for new antibodies and EEEV-143 (PDB 6xob), EEEV-42 (PDB 6mui), EEEV-3 (PDB 6mw9), EEEV-69 (PDB 6mwx), EEEV-58 (PDB 6mwv), EEEV-5 (PDB 6mwc), and LDLRAD3 (PDB 7ffn). (F) Bar graph displaying correlation of binding breadth and epitope properties. Sequence variation was calculated as a BSA-weighted average of normalized entropy. Conformational variability was calculated as a BSA-weighted average RMSD. (G) Epitope sequence and conformational variability plotted for alphavirus antibodies and receptor. Color identifies VLP with which the antibody complex structure was solved. See also Figures S4–S5 and Data S1.