Abstract
The RNA exosome is a conserved molecular machine that processes/degrades numerous coding and non-coding RNAs. The 10-subunit complex is composed of three S1/KH cap subunits (human EXOSC2/3/1; yeast Rrp4/40/Csl4), a lower ring of six PH-like subunits (human EXOSC4/7/8/9/5/6; yeast Rrp41/42/43/45/46/Mtr3), and a singular 3′-5′ exo/endonuclease DIS3/Rrp44. Recently, several disease-linked missense mutations have been identified in structural cap and core RNA exosome genes. In this study, we characterize a rare multiple myeloma patient missense mutation that was identified in the cap subunit gene EXOSC2. This missense mutation results in a single amino acid substitution, p.Met40Thr, in a highly conserved domain of EXOSC2. Structural studies suggest that this Met40 residue makes direct contact with the essential RNA helicase, MTR4, and may help stabilize the critical interaction between the RNA exosome complex and this cofactor. To assess this interaction in vivo, we utilized the Saccharomyces cerevisiae system and modeled the EXOSC2 patient mutation into the orthologous yeast gene RRP4, generating the variant rrp4-M68T. The rrp4-M68T cells show accumulation of certain RNA exosome target RNAs and show sensitivity to drugs that impact RNA processing. We also identified robust negative genetic interactions between rrp4-M68T and specific mtr4 mutants. A complementary biochemical approach revealed that Rrp4 M68T shows decreased interaction with Mtr4, consistent with these genetic results. This study suggests that the EXOSC2 mutation identified in a multiple myeloma patient impacts the function of the RNA exosome and provides functional insight into a critical interface between the RNA exosome and Mtr4.
Keywords: RNA exosome, Mtr4, EXOSC2, Rrp4, RNA processing, multiple myeloma, RNA helicase
This study uses Saccharomyces cerevisiae to model a missense mutation in the RNA exosome gene EXOSC2, which was identified in a patient with multiple myeloma. This analysis of Rrp4, the budding yeast EXCOSC2 ortholog, provides insight into the interaction of Rrp4 with a key RNA exosome cofactor, the RNA helicase Mtr4.
Introduction
The RNA exosome is a highly conserved exo/endonuclease complex that has an essential role in 3′ to 5′ processing and degradation of nearly every species of RNA (Schneider and Tollervey 2013; Zinder and Lima 2017). First identified in Saccharomyces cerevisiae in a screen for ribosomal RNA processing (rrp) mutants (Mitchell et al. 1996; Mitchell et al. 1997), the RNA exosome is essential in all organisms studied thus far (Mitchell et al. 1997; Lorentzen et al. 2007; Hou et al. 2012; Lim et al. 2013; Pefanis et al. 2014). In addition to ribosomal RNA precursors, the RNA exosome processes a variety of small non-coding RNAs (ncRNAs), including small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) (Allmang et al. 1999; Van Hoof et al. 2000; Kilchert et al. 2016; Fasken et al. 2020). The RNA exosome also plays roles in targeting RNA for degradation and decay, including non-functional or aberrant mRNAs and nuclear transcripts that result from pervasive transcription such as cryptic unstable transcripts (CUTs) in budding yeast or promoter upstream transcripts (PROMPTs) in humans (Wyers et al. 2005; Preker et al. 2008; Moore and Proudfoot 2009; Parker 2012; Schneider et al. 2012). The RNA exosome complex is composed of a 9-subunit structural core and a single exo/endonuclease [DIS3/DIS3L (human); Dis3/Rrp44 (budding yeast)]. As shown in Fig. 1a, the 9-subunit structural core is composed of three S1/KH cap subunits (EXOSC1/2/3; Csl4/Rrp4/Rrp40) and a lower ring of six PH-like subunits (EXOSC4/5/6/7/8/9; Rrp41/Rrp46/Mtr3/Rrp42/Rrp43/Rrp45). The nuclear RNA exosome has an additional 3′-5′ exonuclease, EXOSC10/Rrp6, that associates with the complex and aids in nuclear RNA targeting and processing (Briggs et al. 1998; Wasmuth et al. 2014). Structural studies demonstrate that the overall organization of the RNA exosome is conserved (Fig. 1b), suggesting not only evolutionary conservation of the RNA exosome function but structure as well (Makino et al. 2013; Schuller Jan et al. 2018; Weick et al. 2018). The vast array of targets and evolutionary conservation of the complex components indicates a fundamental role of the RNA exosome in several cellular processes, including but not limited to, maintaining genome integrity, translation, and cell differentiation through degradative and surveillance pathways (Ogami et al. 2018).
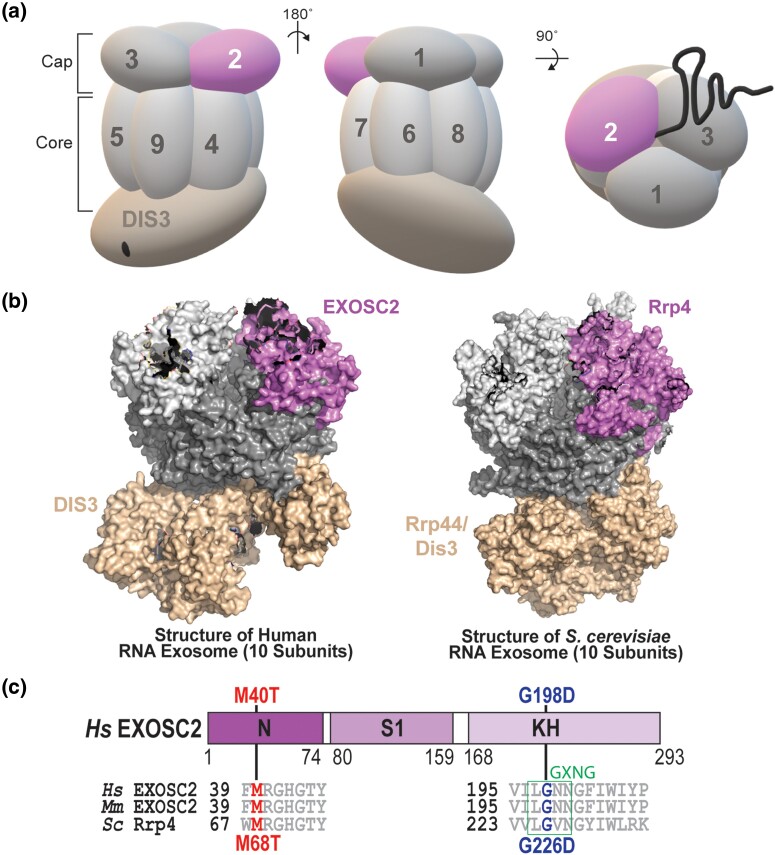
Fig. 1.
Overview of multiple myeloma-linked amino acid substitutions in the human cap subunit EXOSC2 of the RNA exosome. a) The RNA exosome is an evolutionary conserved ribonuclease complex composed of nine structural subunits (EXOSC1-9) and one catalytic subunit (DIS3) that form a “Cap” and “Core” ring-like structure. The three-subunit cap at the top of the complex is composed of EXOSC1/Csl4 (human/S. cerevisiae), EXOSC2/Rrp4, and EXOSC3/Rrp40 (labeled 1–3). The six-subunit core is composed of EXOSC4/Rrp41, EXOSC5/Rrp46, EXOSC6/Mtr3, EXOSC7/Rrp42, EXOSC8/Rrp43, and EXOSC9/Rrp45 (labeled 4–9). The DIS3/Dis3/Rrp44 catalytic subunit is located at the bottom of the complex. Together, the cap and core form a barrel-like structure through which RNA is threaded to the catalytic DIS3/Dis3/Rrp44 subunit. Recent missense mutation in the gene encoding the EXOSC2 cap subunit (pink, labeled "EXOSC2") has been identified in patients presenting with multiple myeloma. b) Structural models of the human nuclear RNA exosome (left) (PDB 6D6R) (Weick et al. 2018) and the S. cerevisiae nuclear RNA exosome (right) (PDB 6FSZ) (Schuller et al. 2018) are depicted with the cap subunit EXOSC2/Rrp4 labeled and colored. c) Domain structure of EXOSC2/Rrp4. This cap subunit is composed of three domains: an N-terminal domain, a putative RNA binding S1 domain, and a C-terminal putative RNA binding KH (K homology) domain. A conserved “GxNG” motif identified in the KH domain is boxed in green (Oddone et al. 2007). The position of the disease-linked amino acid substitutions in human EXOSC2 is depicted above the domain structures. The amino acid substitution (p.Met40Thr) that we report in a multiple myeloma patient is shown in red. An amino acid substitution (p.Gly198Asp) linked to SHRF is shown in blue (Di Donato et al. 2016). Sequence alignments of EXOSC2/Rrp4 orthologs from Homo sapiens (Hs), Mus musculus (Mm), and S. cerevisiae (Sc) below the domain structures show the highly conserved residues altered in disease in red and blue and the conserved sequences flanking these residues in gray.
RNA exosome specificity for a broad set of target transcripts is conferred in part through interactions with cofactor proteins, which aid the RNA exosome in target recognition, RNA unwinding, degradation, and catalysis in both the nucleus and the cytoplasm (Zinder and Lima 2017; Fasken et al. 2020). Many nuclear RNA exosome cofactors were first characterized in budding yeast, including the Rrp6 obligate binding partner Rrp47, the cofactor Mpp6, and the essential 3′ to 5′ DExH box RNA helicase Mtr4 (De La Cruz et al. 1998; Mitchell et al. 2003; Lacava et al. 2005; Vaňáčová et al. 2005; Milligan et al. 2008), with orthologs now identified in the mammalian system (C1D, MPH6, and MTR4/MTREX) (Zinder and Lima 2017). Structural studies of the budding yeast and mammalian RNA exosome reveal that Rrp6/EXOSC10, Rrp47/C1D, and Mpp6/MPH6 interact with the complex through conserved interfaces that form composite sites for interactions with other cofactors such as Mtr4/MTR4/MTREX (Schuch et al. 2014; Falk et al. 2017; Wasmuth et al. 2017; Schuller et al. 2018; Weick et al. 2018). The Mtr4 helicase assists in RNA substrate unwinding and plays a critical role in RNA exosome processing of the 5.8S rRNA precursor (7S rRNA) (De La Cruz et al. 1998; Taylor et al. 2014). Mtr4 also acts as part of larger complexes that aid the RNA exosome in nuclear RNA quality control, including the budding yeast TRAMP (Trf4/5-Air1/2-Mtr4 polyadenylation) complex and the mammalian nuclear exosome targeting (NEXT) complex (Houseley and Tollervey 2008; Houseley and Tollervey 2009; Weir et al. 2010; Lubas et al. 2011; Stuparevic et al. 2013; Falk et al. 2014; Kilchert et al. 2016). Several studies that have dissected the role of Mtr4 in aiding the RNA exosome were performed in the S. cerevisiae system, establishing a number of mtr4 mutations that disrupt specific interactions and functions of the helicase (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996; Weir et al. 2010; Taylor et al. 2014). Thus, genetic model systems are a tractable system to investigate interactions with these nuclear cofactors that impact RNA exosome function and studies in such systems can expand our understanding of the influence that the RNA exosome can exert over various cellular processes and pathways (Cervelli and Galli 2021).
Given the variety of RNA exosome target RNAs and their link to many cellular processes, connections between the RNA exosome and human disease are not surprising. Many different human disease-linked mutations have been identified in genes encoding RNA exosome subunits (Fasken et al. 2020). Mutations in DIS3, which encodes the catalytic component of the RNA exosome in humans (Staals et al. 2010), are the fourth most common single nucleotide variation identified in multiple myeloma (∼10% of all newly diagnosed patients) (Chapman et al. 2011; Lohr et al. 2014). Multiple myeloma, which is a currently incurable cancer of the long-lived antibody-secreting plasma cells of the bone marrow, is the second most common hematologic malignancy accounting for 10–15% of incidence and 20% of deaths related to cancer of the blood and bone marrow (Alexander et al. 2007; Laubach et al. 2011). Multiple myeloma-associated DIS3 mutations disrupt proper RNA degradation and processing in both mammalian cells and budding yeast mutant cells (Tomecki et al. 2014; Weissbach et al. 2015; Boyle et al. 2020). However, additional mechanistic studies are required to understand how mutations in DIS3, and the function of the RNA exosome, could contribute to pathogenesis in multiple myeloma.
Human disease mutations have also been identified in the genes encoding the non-catalytic, structural subunits of the RNA exosome. Clinical studies have linked mutations in EXOSC genes to various, tissue-specific human pathologies comprising a growing family of diseases termed “RNA exosomopathies” (Wan et al. 2012; Boczonadi et al. 2014; Eggens et al. 2014; Di Donato et al. 2016; Burns et al. 2017; Slavotinek et al. 2020; Somashekar et al. 2021). RNA exosomopathy mutations have been found in all three genes that encode the cap subunits (EXOSC1/2/3) and several ring subunit genes (EXOSC5/8/9), with most being missense mutations that result in single amino acid substitutions in highly conserved domains of the subunits. Most RNA exosomopathy diseases are neurological, with mutations in EXOSC1, EXOSC3, EXOSC5, EXOSC8, and EXOSC9 causing forms of cerebellar atrophy/degeneration and neuronopathies (Wan et al. 2012; Boczonadi et al. 2014; Eggens et al. 2014; Burns et al. 2018; Slavotinek et al. 2020; Somashekar et al. 2021). In contrast, patients with RNA exosomopathy mutations in EXOSC2 have a complex syndrome known as SHRF that is characterized by short stature, hearing loss, retinitis pigmentosa, and distinctive facies (OMIM #617763) (Di Donato et al. 2016). In vivo studies characterizing some of these EXOSC RNA exosomopathy mutations in S. cerevisiae and Drosophila melanogaster suggest that these pathogenic substitutions could differentially impact the function of the RNA exosome complex potentially through changes in RNA targeting and cofactor interactions (Fasken et al. 2017; Gillespie et al. 2017; Yang et al. 2019; De Amorim et al. 2020; Morton et al. 2020; Slavotinek et al. 2020; Sterrett et al. 2021). Modeling these pathogenic amino acid substitutions in the budding yeast RNA exosome is an invaluable tool as several RNA exosomopathies have a small patient population, making analysis with patient tissue samples challenging. Therefore, by utilizing the budding yeast system, we can begin elucidating the functional and molecular consequences resulting from human disease mutations in RNA exosome genes (Cervelli and Galli 2021).
In this study, we report several missense mutations in genes encoding structural subunits of the human RNA exosome that were identified from analysis of multiple myeloma patients and characterize the functional consequences of one of these mutations. For this analysis, we surveyed the ongoing longitudinal Multiple Myeloma Research Foundation (MMRF) study “Relating Clinical Outcomes in Multiple Myeloma to Personal Assessment of Genetic Profile” (CoMMpass) (ClinicalTrials.gov Identifier NCT01454297) to identify mutations in structural RNA exosome genes within multiple myeloma patients (Barwick et al. 2019). We focused on characterizing EXOSC2 M40T, a missense mutation that encodes an amino acid substitution EXOSC2 p.Met40Thr (M40T) in a highly conserved region of this cap subunit that interacts with the RNA helicase MTR4. To assess the effects of this amino acid substitution in EXOSC2 on the function of the RNA exosome, we utilized the budding yeast model system and generated a variant of the S. cerevisiae EXOSC2 ortholog, Rrp4, which models the patient EXOSC2 M40T substitution, Rrp4 M68T. As a comparative control within our studies, we included the Rrp4 G226D variant, which models a SHRF-linked pathogenic amino acid substitution in EXOSC2 p.Gly198Asp (Sterrett et al. 2021). The rrp4-G226D cells, corresponding to the SHRF EXOSC2 exosomopathy mutation, have defects in RNA exosome function and are the only other budding yeast model of a disease-linked EXOSC2 mutation (Sterrett et al. 2021). Our results show that the rrp4-M68T gene variant can replace the function of the essential RRP4 gene. The rrp4-M68T and rrp4-G226D mutants show similar increases in specific RNA exosome target transcripts, suggesting shared defects in RNA processing. However, the rrp4-M68T mutant exhibits distinct negative genetic interactions with RNA exosome cofactor mutants, particularly mtr4 mutants. A binding assay provides evidence that the M68T substitution impairs the interaction of Rrp4 with Mtr4. Combined, our results suggest that the Rrp4 M68T amino acid substitution, which models the multiple myeloma-associated substitution EXOSC2 M40T, alters RNA exosome function by impacting the essential interaction between the complex and Mtr4. These data are the first in vivo characterization of this isolated multiple myeloma-associated mutation and give insight into the critical and conserved interactions between the RNA exosome and its cofactors.
Materials and methods
Media and chemicals
All media were prepared by standard procedures (Adams et al. 1997). Unless stated otherwise, all chemicals were acquired from Fisher Scientific (Pittsburgh, PA), Sigma-Aldrich (St. Louis, MO), or United States Biological (Swampscott, MA).
In silico protein structure predictions
The mCSM-PPI2 platform (Rodrigues et al. 2019) and the PyMOL viewer (The PyMOL Molecular Graphics System, version 2.0, Schrödinger, LLC) (PyMOL) were used for structural modeling. Platforms were used with the cryo-EM structure (PDB 6D6Q) of the human nuclear RNA exosome at 3.45Å resolution (Weick et al. 2018) and the X-ray diffraction structure (PDB 6FSZ) of the budding yeast nuclear RNA exosome at 4.60Å (Schuller Jan et al. 2018). The ConSurf server (Ashkenazy et al. 2010; Celniker et al. 2013; Ashkenazy et al. 2016) was used to assess the evolutionary conservation of the structure of both EXOSC2 and Rrp4.
Saccharomyces cerevisiae strains and plasmids
All DNA manipulations were performed according to standard procedures (Sambrook et al. 1989). Saccharomyces cerevisiae strains and plasmids used in this study are listed in Supplementary Table 1. The rrp4Δ (yAV1103), rrp4Δ mpp6Δ (ACY2471), and rrp4Δ rrp47Δ (ACY2474) strains have been previously described (Schaeffer et al. 2009; Losh 2018; Sterrett et al. 2021). The RRP45-TAP (ACY2789) strain was obtained from Horizon Discovery Biosciences Limited and was previously described (Ghaemmaghami et al. 2003). The mtr4Δ (ACY2532) strain was constructed by deletion of the genomic MTR4 ORF in a wild-type (W303) strain harboring a (MTR4, RRP4, and URA3) (pAC3714) maintenance plasmid by homologous recombination using MTR4-UTR natMX4. This mtr4Δ (ACY2532) strain was then used for consecutive deletion of the genomic RRP4 ORF to generate the rrp4Δmtr4Δ (ACY2536) strain as previously described (Sterrett et al. 2021). Construction of the untagged RRP4 and rrp4-G226D plasmids (pAC3656 and pAC3659) and the 2x-Myc-tagged RRP4 and rrp4-G226D plasmids (pAC3669 and pAC3672) that contain native 3′ UTRs was reported previously (Sterrett et al. 2021). The rrp4-M68T LEU2 CEN6 (pAC4206) and rrp4-M68T-2xMyc LEU2 CEN6 (pAC4207) plasmids were generated by site-directed mutagenesis of the RRP4 (pAC3656) or RRP4-2xMyc (pAC3669) plasmids using oligonucleotides containing the M68T missense mutation (Fwd 5′GAAAATACGTACCGTGACCT CTCGTCCAGATAGGGTCATCAGTGACC 3′, Rev 5′GGTCACTGATGACCCTATCTGGACGA GAGGTCACGGTACGTATTTTC 3′) and the QuikChange II Site-Directed Mutagenesis Kit (Agilent). The mtr4-F7A-F10A (pAC4099) plasmid was generated as described previously (Sterrett et al. 2021). Similarly, the other mtr4 mutant plasmids were constructed by site-directed mutagenesis of the MTR4 HIS CEN6 plasmid (pAC4096) with the QuikChange II Site-Directed Mutagenesis Kit (Agilent) and oligonucleotides containing the corresponding missense mutations. The oligonucleotides used to generate the mtr4-1 plasmid (pAC4103) contain the C942Y missense mutation (Fwd 5′CAAGCAGCAGCATTATTATCATACTTTG CATTCCAAGAACGCTG 3′, Rev 5′CAGCGTTCTTGGAATGCAAAGTATGATAATAATG CTGCTGCTTG 3′). The oligonucleotides used to generate the mtr4-R349E-N352E plasmid (pAC4100) contain the R349E and N352E missense mutations (Falk et al. 2014) (Fwd 5′ GGTTGACGAAAAAAG TACCTTCGAAGAGGAAGAATTCCAAAAAGCAATGGCGTCC 3′, Rev 5′ GGACGCCATTGC TTTTTGGAATTCTTCCTCTTCGAAGGTACTTTTTTCGTCAACC 3′). The oligonucleotides used to generate the mtr4-R1030A plasmid (pAC4104) contain the R1030A missense mutation (Taylor et al. 2014) (Fwd 5′CGTTGATCAGAATGTTCAAGGCATTAGAGGAATTGGTGAAGG 3′, Rev 5′CCTTCA CCAATTCCTCTAATGCCTTGAACATTCTGATCAACG 3′) and the oligonucleotides used to generate the mtr4-E1033W plasmid (pAC4105) contain the E1033W missense mutation (Taylor et al. 2014) (Fwd 5′GAATGTTCAAGAGATTAGAGTGGTTGGTGAAGGAGCTGGTAGAC 3′, Rev 5′GTCTACCAGCTC CTTCACCAACCACTCTAATCTCTTGAACATTC). Plasmids were confirmed through DNA sequencing.
Saccharomyces cerevisiae transformations and growth assays
All S. cerevisiae transformations were conducted following the standard lithium acetate (LiOAc) protocol (Da et al. 2000). Strains were grown in liquid YEPD (1% yeast extract, 2% peptone, 2% dextrose, in distilled water) in a rotating shaker at 30°C overnight to saturation. Cultures were normalized to a concentration of OD600 = 0.33 in 10 mL YEPD and then incubated at 30°C for 3–8 hours depending on the severity of their growth defect. Cells were washed and resuspended to a concentration of 2 × 109 cells/mL using TE/LiOAc. Single-stranded carrier DNA (5 μL; 10 mg/mL), PEG/TE/LiOAc (300 μL), and, depending on reaction purpose, desired PCR product DNA or plasmid DNA, were added to cells. The mixture was incubated at 30°C in a shaker for 30 minutes. Following this incubation, DMSO (35 μL) was added and the cells were heat shocked for 15 minutes at 42°C, washed, and plated onto selective media.
Standard plasmid shuffle assays were performed to assess the in vivo function of the rrp4 variants as well as genetic interaction with RNA exosome cofactor mutants. The rrp4Δ (yAV1103) cells containing a RRP4 URA3 maintenance plasmid and transformed with vector (pRS315) and transformed with RRP4 (pAC3656), rrp4-G226D (pAC3659), rrp4-M68T (pAC4206), RRP4-2xMyc (pAC3669), or rrp4-M68T-2xMyc (pAC4207) plasmid were grown on Ura− Leu− minimal media control plates, which select for cells that contain both the RRP4 URA3 maintenance plasmid as well as the RRP4/rrp4 LEU2 plasmid, and 5-FOA Leu− minimal media plates, which select for cells that lack the RRP4 URA3 maintenance plasmid and contain only the RRP4/rrp4 LEU2 plasmid. The plates were incubated at 30°C for 2–3 days and single colonies from the 5-FOA Leu− minimal media plates were selected in quadruplicate and streaked onto selective Leu− minimal media plates. The cells containing only the RRP4/rrp4 LEU2 plasmid are referred to as RRP4, rrp4-G226D, or rrp4-M68T cells. A similar strategy was used to generate mtr4Δ (ACY2532) cells that contain only the MTR4 (pAC4096) or mtr4-1 (pAC4103) HIS3, CEN6 plasmid. The mtr4Δ cells transformed with MTR4 or mtr4-1 were grown overnight and serially diluted and spotted onto Ura− His− minimal media plates and 5-FOA minimal media plates, which select for cells that lack the URA3 maintenance plasmid and contain only the MTR4/mtr4 HIS3 plasmid. Single colonies of cells containing only MTR4 or mtr4-1 HIS3 plasmid were collected in quadruplicate and are referred to as MTR4 or as mtr4-1 cells.
The in vivo function of the rrp4-M68T variant was assessed in growth assays on solid media and in liquid culture. For growth on solid media, rrp4Δ (yAV1103) cells containing only RRP4 (pAC3656), rrp4-G226D (pAC3659), or rrp4-M68T (pAC4206) were grown in 2 mL Leu− minimal media overnight at 30°C to saturation. Cell concentrations were normalized to an OD600 = 1, and samples were serially diluted in 10-fold dilutions and spotted onto Leu− minimal media plates, Leu− minimal media plates containing 25 µM fluorouracil (5-FU), YEPD plates, or YEPD plates containing 3% formamide, 150 mM hydroxyurea, or 5 µg/mL phleomycin. Plates were grown at 25°C, 30°C, and 37°C for 2–3 days. For growth in liquid culture, cells were grown in 2 mL Leu− minimal media overnight at 30°C to saturation and diluted to an OD600 = 0.05 in Leu− minimal media in a 24-well plate and growth at 37°C was monitored and recorded at OD600 in a BioTek SynergyMx microplate reader with Gen5 v2.04 software over 36 hr. For these liquid growth assays, the cells incubate in the microplate reader for many hours before their density is within the dynamic range of the machine to record the doubling times. For the results shown, each sample was performed in at least three independent biological replicates with three technical replicates for each biological sample. Doubling times were calculated using GraphPad Prism version 9.3.1 for Windows (www.graphpad.com), GraphPad Software, San Diego, California, USA.
Immunoblotting
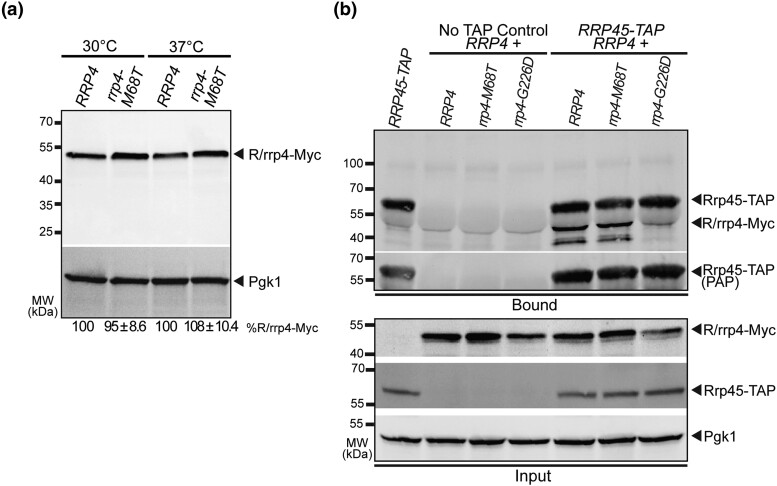
To analyze protein expression levels of C-terminally Myc-tagged Rrp4 and Rrp4 M68T, rrp4Δ (yAV1103) cells expressing only Rrp4-2xMyc (pAC3669) or Rrp4-M68T-2xMyc (pAC4207) were incubated in 2 mL of Leu− minimal medium at 30°C and grown to saturation overnight. The 10 mL cultures with an OD600 = 0.2 were prepared and incubated at 30°C or 37°C for 5 hr. Yeast cell pellets were collected by centrifugation and transferred to 2 mL screw-cap tubes. Cell pellets were flash frozen with liquid nitrogen and stored at −20°C. Yeast cell lysate was prepared by resuspending pellets in 0.3 mL of RIPA-2 Buffer (50 mM Tris-HCl, pH 8; 150 mM NaCl; 0.5% sodium deoxycholate; 1% NP40; 0.1% SDS) supplemented with protease inhibitors [1 mM PMSF; 3 ng/mL PLAC (pepstatin A, leupeptin, aprotinin, and chymostatin)], followed by addition of 300 µL glass beads. Lysates were placed in a Mini Bead Beater 16 Cell Disrupter (Biospec) for 6 × 1 min at 25°C with ice submersion intervals of 1 minute between rounds and then centrifuged at 4°C at 12,000 RPM for 10 min. Protein lysate concentration was determined by Pierce BCA Protein Assay Kit (Life Technologies). Whole cell lysate protein samples (40 µg) in reducing sample buffer (50 mM Tris HCl, pH 6.8; 100 mM DTT; 2% SDS; 0.1% Bromophenol Blue; 10% Glycerol) were resolved on criterion 4–20% gradient denaturing gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad) and Myc-tagged Rrp4 proteins were detected with anti-Myc monoclonal antibody 9B11 (1 : 2,000; Cell Signaling). The 3-phosphoglycerate kinase (Pgk1) protein was detected using anti-Pgk1 monoclonal antibody (1 : 30,000; Invitrogen) as a loading control. For quantitation, ImageJ v1.4 software (National Institutes of Health, MD; http://rsb.info.nih.gov/ij/) was used to measure protein band areas and intensities. Protein percentages relative to Pgk1 were calculated using GraphPad Prism version 9.3.1 for Windows (www.graphpad.com), GraphPad Software, San Diego, California, USA.
Coimmunoprecipitations
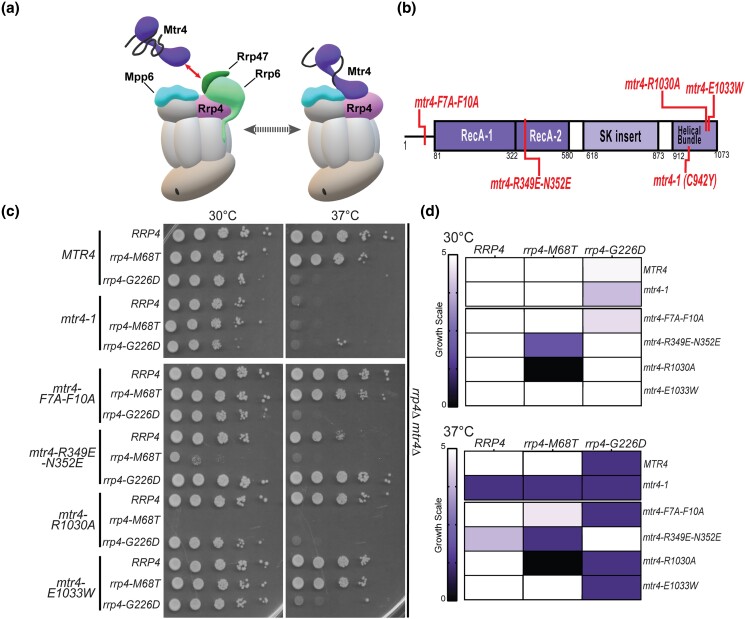
To assess association of Rrp4 M68T with the RNA exosome complex, we utilized RRP45-TAP (ACY2789) cells expressing RRP4-2xMyc (pAC3669), rrp4-G226D-2xMyc (pAC3672), or rrp4-M68T-2xMyc (pAC4207) and immunoprecipitated Rrp45-TAP using the IgG Sepharose beads as previously described (Sterrett et al. 2021). Briefly, cell samples were grown in 2 mL Leu− minimal media overnight at 30°C to saturation and 10–20 mL cultures with an OD600 = 0.2 were prepared and grown at 30°C for 5 hr. Yeast cell lysates were prepared by resuspending cell pellets in 0.5–0.75 mL of IPP150 Buffer (10 mM Tris-HCl, pH 8; 150 mM NaCl; 0.1% NP40, PMSF) supplemented with protease inhibitors [1 mM PMSF; Pierce Protease Inhibitors (Thermo Fisher Scientific)] and 300 µL of glass beads. Cells were disrupted in a Mini Bead Beater 16 Cell Disrupter (Biospec) for 4–5 × 1 min at 25°C with 1 min on ice between repetitions. Crude lysate was transferred to a chilled microcentrifuge tube and remaining beads were washed with an additional 150 µL of IPP150 Buffer. Lysate was then cleared by centrifugation at 16,000 × g for 10 min at 4°C. Protein lysate concentration was determined by Pierce BCA Protein Assay Kit (Life Technologies). For input samples, 40 µg of cleared lysate was collected and frozen at −20°C. For coimmunoprecipitations, 1 mg of cleared lysate in IPP150 Buffer was prepared, 30 µL of a 1 : 1 bead slurry of IgG Sepharose 6 Fast Flow Beads (GE Healthcare) was added, and samples were incubated at 4°C overnight with mixing. After overnight incubation, beads were washed three times in 1 mL IPP150 Buffer for 5 min each (IgG Sepharose beads). Whole cell lysate input samples (40 µg) and total bound samples in reducing sample buffer were boiled for 5 min at 100°C, resolved on 4–20% Criterion TGX precast polyacrylamide gels (Bio-Rad), transferred to nitrocellulose membranes (Bio-Rad). Levels of associated Rrp4-Myc proteins with the Rrp45-TAP-tagged subunit were detected by immunoblotting. Myc-tagged Rrp4 proteins were detected with mouse anti-Myc monoclonal antibody 9B11 (1 : 2,000; Cell Signaling). TAP-tagged Rrp45 protein was detected with peroxidase–antiperoxidase (PAP) soluble complex antibody produced in rabbit (1 : 5,000, Sigma-Aldrich). The 3-phosphoglycerate kinase (Pgk1) protein was detected using anti-Pgk1 monoclonal antibody (1 : 30,000; Invitrogen) as a loading control.
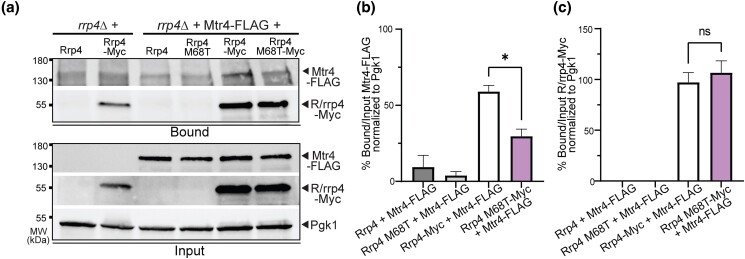
To assess the association of Mtr4 with Rrp4 M68T, we utilized rrp4Δ cells expressing Rrp4-2xMyc (pAC3669) or Rrp4-M68T-2xMyc (pAC4207). These cells were transformed with either an empty vector plasmid (pAC1) or a plasmid to express 2x-FLAG-tagged Mtr4 (pAC3719). Briefly, cell samples were grown in 2 mL His−Leu− minimal media overnight at 30°C to saturation and 40 mL cultures with an OD600 = 0.2 were prepared and grown at 30°C for 4 hr. Yeast cell lysates were prepared as described above. Cleared protein lysate concentration was determined by Pierce BCA Protein Assay Kit (Life Technologies). For input samples, 50 µg of cleared lysate was collected and frozen at −20°C. For coimmunoprecipitations, 1.75 mg of cleared lysate in IPP150 Buffer was prepared, 15 µL of a 1 : 1 bead slurry of Pierce Anti-c-Myc Magnetic Beads (ThermoFisher) was added, and samples were incubated at 4°C overnight with mixing. After overnight incubation, beads were washed three times in 1 mL IPP150 Buffer for 15 sec each. Whole cell lysate input samples (50 µg) and total bound samples in reducing sample buffer were boiled for 10 min at 100°C, resolved on 4–20% Criterion TGX precast polyacrylamide gels (Bio-Rad), and transferred to nitrocellulose membranes (Bio-Rad). Levels of Mtr4-FLAG protein associated with Rrp4-Myc or Rrp4 M68T-Myc-tagged subunit were detected by immunoblotting. Myc-tagged Rrp4 proteins were detected with mouse anti-Myc monoclonal antibody 9B11 (1 : 2,000; Cell Signaling). FLAG-tagged Mtr4 protein was detected with anti-FLAG monoclonal antibody 9A3 (1 : 1,000; Cell Signaling). The 3-phosphoglycerate kinase (Pgk1) protein was detected using anti-Pgk1 monoclonal antibody (1 : 30,000; Invitrogen) as a loading control. A peroxidase AffiniPure Goat Anti-Mouse IgG, light chain specific secondary was used to detect bound proteins < 70 kDa in size. Quantitation was performed first by standardizing detected FLAG-tagged and Myc-tagged protein levels to Pgk1 levels. Then, protein levels from bound samples were normalized to unbound levels to generate a value for %Bound/Input normalized to Pgk1.
Genetic interaction analysis
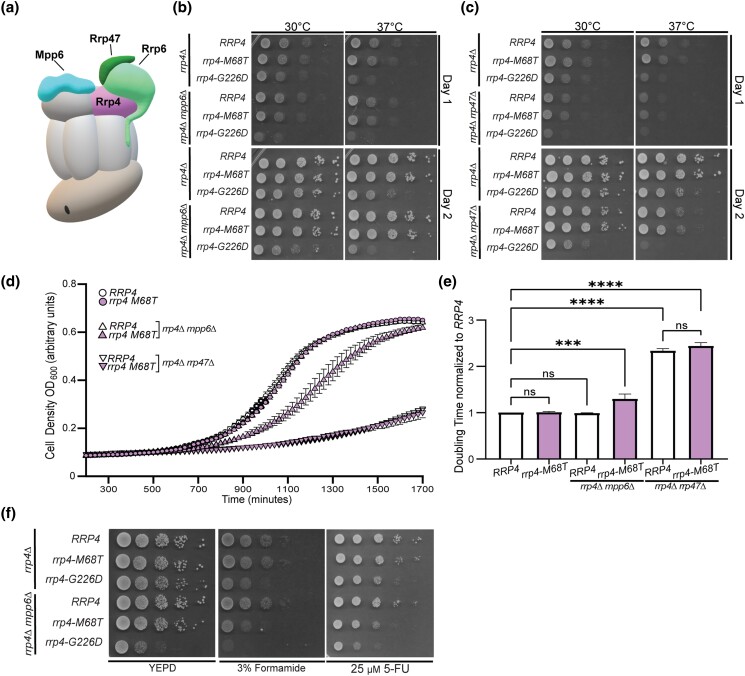
To test genetic interactions between rrp4-M68T and RNA exosome cofactor/subunit deletion mutants, rrp4Δ mpp6Δ (ACY2471) and rrp4Δ rrp47Δ (ACY2474) cells containing only RRP4 (pAC3656), rrp4-G226D (pAC3659), or rrp4-M68T (pAC4206) were grown in 2 mL Leu− minimal media overnight at 30°C to saturation, serially diluted, and spotted on Leu− minimal media plates. The plates were incubated at 30°C or 37°C for 3 days. Cells were also grown in liquid culture as described in S. cerevisiae transformation and growth assay method. The rrp4Δ mpp6Δ (ACY2471) cells containing only RRP4 (pAC3656), rrp4-G226D (pAC3659), or rrp4-M68T (pAC4206) were further assayed by being serially spotted onto Leu− minimal media plates containing 25 µM 5-FU, YEPD plates, or YEPD plates containing 3% formamide.
To test for genetic interactions between rrp4-M68T and mtr4 mutants, mtr4-F7A-F10A, mtr4-1, mtr4-R1030A, and mtr4-E1033W, rrp4Δ mtr4Δ (ACY2536) cells containing the (MTR4, RRP4, and URA3) (pAC3714) maintenance plasmid were transformed with RRP4 (pAC3656), rrp4-G226D (pAC3659), or rrp4-M68T (pAC4206) LEU2 plasmid and selected on Ura−Leu− minimal media plates. Transformed cells containing both the URA3 maintenance plasmid and the RRP4/rrp4 variant plasmid were subsequently transformed with MTR4 (pAC4096), mtr4-F7A-F10A (pAC4099), mtr4-1 (pAC4103), mtr4-R1030A (pAC4104), or mtr4-E1033W (pAC4105) HIS3 plasmid and selected on Ura−Leu−His− minimal media plates. The transformed cells were then streaked to onto 5-FOA Leu− His− plates to select for cells that did not contain the URA3 maintenance plasmid. The resulting rrp4Δ mtr4Δ cells containing only RRP4, rrp4-G226D, or rrp4-M68T LEU2 plasmid and MTR4 or mtr4 variant HIS3 plasmid were grown in 2 mL Leu− His− minimal media overnight at 30°C to saturation, serially diluted, and spotted on Leu− His− minimal media plates. The plates were incubated at 30°C and 37°C for 3 days. Cell growth was quantified on a scale from 0 to 5 across triplicate assays, with a score of “0” representing no growth detected and a score of “5” representing full growth across all dilutions analyzed. Scores were averaged and displayed as a heatmap using GraphPad Prism version 9.3.1 for Windows (www.graphpad.com), GraphPad Software, San Diego, California, USA.
Total RNA isolation
Total RNA from RRP4, rrp4-G226D, rrp4-M68T, MTR4, or mtr4-1 cells was isolated using the MasterPure Yeast RNA Purification Kit (Epicentre, Lucigen). Cells were incubated in 2 mL of Leu− minimal medium at 30°C and grown to saturation overnight. Cultures were diluted in 10 mL to an OD600 = 0.2 and further incubated at 37°C for 5 hours. Cells were pelleted by centrifugation, transferred to RNase-free microcentrifuge tubes, and flash frozen with liquid nitrogen. Frozen cell pellets were stored at −80°C. RNA isolation was performed according to the MasterPure Yeast RNA Purification Kit (Epicentre, Lucigen) manufacturer's protocol. Total RNA was resuspended in 50 µL DEPC-treated water and stored at −80°C.
RT-qPCR
All oligonucleotides used in this study are shown in Supplementary Table 2. For analysis of steady-state RNA levels using quantitative PCR, three independent biological replicates of RRP4, rrp4-G226D, rrp4-M68T, mtr4-1, and MTR4 cells were grown in 2 mL Leu− or His− minimal media overnight at 30°C. Cultures (10 mL) with an OD600 = 0.2 were prepared from the saturated cultures and cells were grown at 37°C for 5 hr. Total RNA was isolated from cell pellets as described and 1 μg of total RNA was reverse transcribed to first-strand cDNA using the M-MLV Reverse Transcriptase (Invitrogen) according to the manufacturer's protocol. Quantitative PCR was performed on technical triplicates of cDNA (10 ng) from three independent biological replicates using gene-specific primers (0.5 mM; Supplementary Table 2), QuantiTect SYBR Green PCR master mix (Qiagen) on a StepOnePlus Real-Time PCR machine (Applied Biosystems; Tanneal = 55°C, 44 cycles). ALG9 or PGK1 was used as an internal control. The mean RNA levels were calculated by the ΔΔCt method (Livak and Schmittgen 2001). Statistical analysis comparing the control cells (RRP4 or MTR4) and the mutant cells (rrp4 or mtr4-1) was performed by t-test (α < 0.05) using GraphPad Prism version 9.3.1 for Windows (www.graphpad.com), GraphPad Software, San Diego, California, USA.
Results
EXOSC2 p.Met40Thr substitution is located within a conserved region of the cap subunit that interacts with the essential helicase MTR4
Mutations in the gene DIS3, which encodes the catalytic component of the RNA exosome, are commonly found in patients diagnosed with multiple myeloma (Chapman et al. 2011; Lohr et al. 2014), suggesting a link between RNA exosome function and disease pathology. We therefore considered whether mutations in the other components of the RNA exosome would be found in multiple myeloma patients. Missense mutations in EXOSC genes, which encode the structural subunits of the RNA exosome, were identified in multiple myeloma patients through interrogating the ongoing longitudinal MMRF study “CoMMpass” (ClinicalTrials.gov Identifier NCT01454297). A total of 1,154 newly diagnosed multiple myeloma patients were enrolled in CoMMpass and profiled by genomic testing and tissue sampling throughout treatment. The molecular profiling collected through CoMMpass reveals several rare missense mutations within EXOSC genes (Supplementary Fig. 1). One patient missense mutation identified within exon 1 of EXOSC2 encodes EXOSC2 p.Met40Thr (M40T), which is located in a highly conserved region of the N-terminal domain of EXOSC2 (Fig. 1c). Notably, EXOSC2 Met40 lies within a key binding interface between the human RNA exosome and the RNA helicase MTR4 (Weick et al. 2018).
The patient with the EXOSC2 M40T mutation also had chromosomal aberrations including a chromosomal translocation t(11; 14) and hyperdiploidy disease. The chromosomal translocation t(11; 14) is an IgH translocation, which is an initiating event that occurs frequently in multiple myeloma (∼15–20% of patients) (Bergsagel et al. 1997). From the CoMMpass dataset, we determined that the variant allele frequency is 0.2266; however, the copy number of the chromosome 9 EXOSC2 locus is 2.6, suggesting that this EXOSC2 allele is found on the extra copy of ch9 that is present in over half the cells in the patient. Based on these findings, we conclude that this hyperdiploidy of chromosome 9 occurred after the t(11; 14) translocation event and that the EXOSC2 mutation either cooccurred with the chromosomal gain or shortly after.
To explore how EXOSC2 M40T could alter the function of the RNA exosome complex, we modeled the EXOSC2 M40T amino acid substitution using a recent structure of the human RNA exosome in complex with the essential RNA helicase MTR4 (Weick et al. 2018). MTR4 makes several direct contacts with the RNA exosome, forming a binding interface with a total surface area of 1,440Å2 (Weick et al. 2018). Among the direct contacts between MTR4 and the RNA exosome complex, the N-terminal domain of EXOSC2 interacts with MTR4 through an aliphatic surface that includes Met40. As shown in Fig. 2a, EXOSC2 Met40 engages with a hydrophobic pocket of MTR4 including Ile1014. An amino acid substitution of Thr40, while unlikely to disrupt the aliphatic surface, could disrupt the hydrophobic interaction at this contact given the polar, shortened side chain of threonine (Fig. 2a). The EXOSC2 M40T substitution could therefore destabilize the interface between the N-terminal domain of EXOSC2 and MTR4. We also modeled the amino acid substitution in the budding yeast EXOSC2 ortholog, Rrp4, using a recent structure of the S. cerevisiae RNA exosome (Schuller et al. 2018). As shown in Fig. 2b, the budding yeast RNA exosome in complex with Mtr4 shows structural similarities to the human complex, with Rrp4 interacting directly with Mtr4. Rrp4 Met68, corresponding to the EXOSC2 Met40 residue, engages with the helicase directly through hydrophobic interactions at the binding interface. This binding interface of yeast Mtr4 is part of a large hydrophobic pocket which includes Phe924 and Ile923 (not labeled), similar to hydrophobic pocket in the human MTR4 helicase. Introduction of Thr40 would most likely disrupt this contract, similar to our predictions for the M40T substitution in EXOSC2. Furthermore, the region surrounding Rrp4 Met68 is structurally synonymous to the aliphatic surface surrounding EXOSC2 Met40, allowing for us to assess the effects of the EXOSC2 M40T amino acid substitution at the conserved interface within the yeast system.
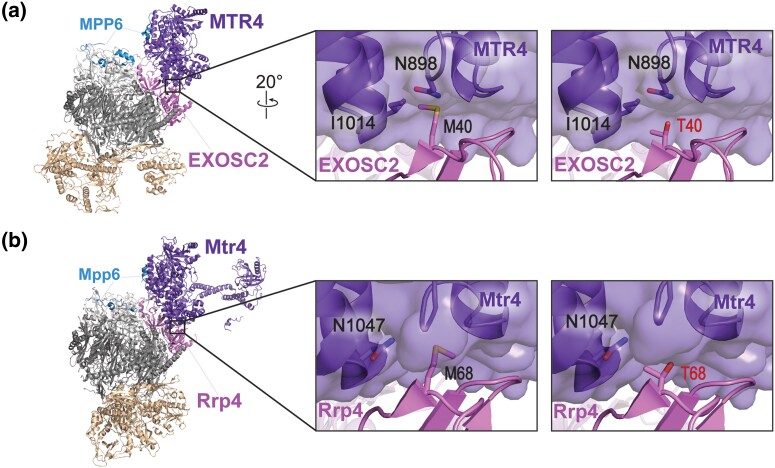
Fig. 2.
Modeling the multiple myeloma EXOSC2 M40T amino acid substitution in the human EXOSC2 cap subunit and the S. cerevisiae ortholog Rrp4. a) Structural modeling of the human EXOSC2 p.Met40Thr (M40T) amino acid substitution identified in a patient with multiple myeloma (PDB 6D6R) (Weick et al. 2018). The full structure of the human RNA exosome with the associated cofactor MTR4 (purple, labeled "MTR4") is depicted with a zoomed-in representation of the interface between EXOSC2 (pink, labeled "EXOSC2") and MTR4. Modeling of the native EXOSC2 Met40 (M40) residue (left) or the multiple myeloma-associated EXOSC2 Thr40 (T40) residue (right) is shown. The EXOSC2 Met40 residue is located in the N-terminal domain of EXOSC2, within a conserved aliphatic interface with MTR4. EXOSC2 Met40 and MTR4 associate through hydrophobic interactions, which include contacts with MTR4 Ile1014 (I1014). b) Structural modeling of the budding yeast Rrp4 Met68Thr (M68T) amino acid change, corresponding to EXOSC2 p.Met40Thr, in the budding yeast RNA exosome (PDB 6FSZ) (Schuller et al. 2018). The full structure of the budding yeast RNA exosome complex with the associated MTR4 ortholog, Mtr4 (purple, labeled "Mtr4"), is depicted on the left. A zoomed-in representation of the interface between Rrp4 (pink, labeled "Rrp4") and Mtr4 are shown, modeling the native Rrp4 Met68 (M68) residue (left) or the modeled multiple myeloma-associated substitution Rrp4 Thr68 (T68) residue (right). The Rrp4 Met68 residue is conserved between human and yeast and is located in the N-terminal domain of Rrp4. Similar to EXOSC2 Met40, Rrp4 Met68 associates with the helicase Mtr4 through primarily hydrophobic interactions, including contacts with several glycine residues in a neighboring loop of Mtr4. Parts of the human nuclear cofactor protein MPP6/MPH6 (blue, labeled "MPP6") and the budding yeast ortholog Mpp6 (blue, labeled "Mpp6") are also resolved in the structures shown in a) and b). Both MPP6/MPH6 and Mpp6 aid in stabilizing the interaction of the RNA helicase with the RNA exosome in addition to the direct interface made between EXOSC2 and MTR4 in humans or Rrp4 and Mtr4 in budding yeast (Falk et al. 2017; Wasmuth et al. 2017).
We further explored the conservation of the binding interface of human EXOSC2 as compared to budding yeast Rrp4 using the bioinformatics tool ConSurf (Supplementary Fig. 2). The ConSurf server estimates the evolutionary conservation of amino acids of a protein based on phylogenetic trees between homologous sequences, providing conservation rates for each residue that reflect both functional and structural importance (Ashkenazy et al. 2010; Celniker et al. 2013; Ashkenazy et al. 2016). Consistent with the sequence alignment (Fig. 1c) and structural modeling, this tool predicts high conservation for both EXOSC2 Met40 and Rrp4 Met68 and surrounding residues (Supplementary Fig. 2). Additionally, ConSurf estimates high conservation rates at each site of contact between EXOSC2 and Rrp4 with the helicase MTR/Mtr4, further supporting the evolutionary importance of this interaction.
Saccharomyces cerevisiae rrp4-M68T mutant cells that model the multiple myeloma EXOSC2 M40T variant show sensitivity on drugs that impact RNA metabolism
To assess the functional consequences of the EXOSC2 M40T amino acid substitution, we generated the corresponding amino change in the S. cerevisiae ortholog Rrp4, M68T (Fig. 1c). We first performed a plasmid shuffle growth assay in which cells deleted for the genomic copy of RRP4 are transformed with plasmids containing different rrp4 alleles (see Materials and Methods). This approach ensures that the background for all variants that are compared to one another is identical (Sikorski and Hieter 1989). This growth assay reveals that the rrp4-M68T allele can replace the essential RRP4 gene as the rrp4-M68T cells grow similarly to control cells expressing wild-type RRP4 at all temperatures examined (Fig. 3a). As a comparison, we included cells expressing the rrp4-G226D allele as the sole copy of the essential RRP4 gene. The rrp4-G226D mutant allele models a known SHRF pathogenic amino acid change that has been shown to cause impaired RNA exosome function in vivo (Sterrett et al. 2021). As previously reported, cells expressing only rrp4-G226D show impaired growth at 37°C (Fig. 3a). Furthermore, we assessed the growth of both the rrp4-M68T and rrp4-G226D mutant cells using a liquid growth assay and quantified doubling times (Fig. 3b, 3c). These data confirm that the growth of rrp4-M68T cells does not differ significantly from wild-type RRP4 cells.
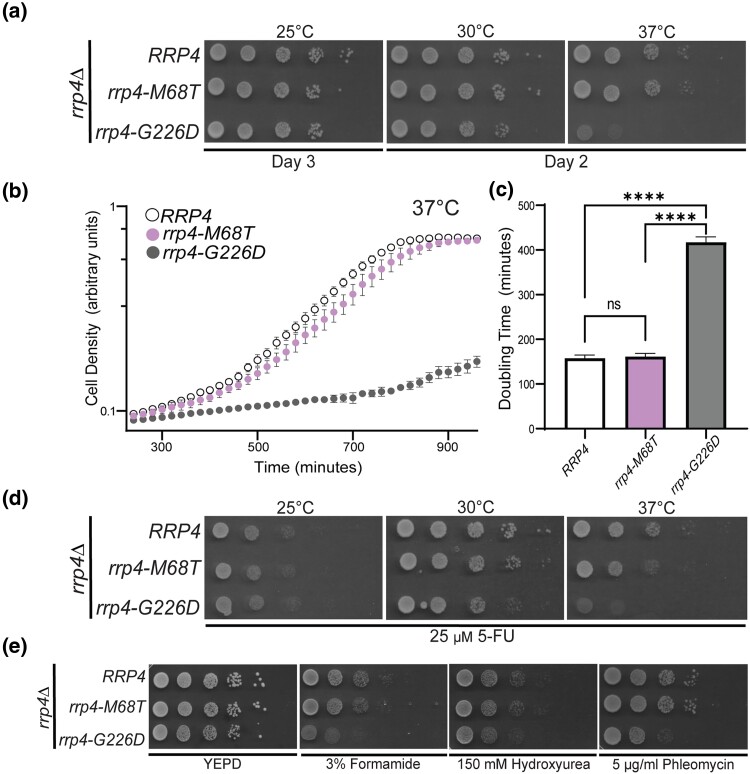
Fig. 3.
S. cerevisiae rrp4-M68T mutant cells that model the EXOSC2 M40T variant identified in multiple myeloma patients show impaired function on drugs that impact RNA processing. S. cerevisiae cells expressing Rrp4 variants that model the multiple myeloma amino acid change or, as a control, the previously characterized (Sterrett et al. 2021) SHRF-linked amino acid change found in EXOSC2 were generated as described in Materials and Methods. a–b) The rrp4Δ cells expressing only RRP4 or mutant rrp4 were serially diluted, spotted onto solid selective media grown at the indicated temperatures, or grown in liquid media at 37°C with optical density measurement used to assess cell density over time. The doubling time of these cells grown in liquid media is quantified and graphed in c). d) The rrp4Δ cells expressing either RRP4 or rrp4-M68T were serially diluted, spotted onto solid selective media containing 25 µM fluorouracil (5-FU), and grown at the indicated temperatures. Images shown are after 2 days of growth. e) The rrp4Δ cells expressing only RRP4 or rrp4-M68T were serially diluted and spotted onto solid YEPD media containing 3% formamide, 150 mM hydroxyurea, or 5 µg/ml phleomycin and grown at 30°C. Images shown are after two days of growth. In all assays performed, rrp4-G226D cells, previously reported to be severely impaired at 37°C, were included as a control (Sterrett et al. 2021). Data shown are representative of three independent experiments (n = 3).
To explore whether the rrp4-M68T mutation sensitizes cells to altered RNA processing, we tested for growth defects when cells are grown on media containing 5-fluorouracil (5-FU) (Giaever et al. 2004; Hoskins and Scott Butler 2007) (Fig. 3d). The rrp4-M68T cells show a slight growth defect compared to wild-type RRP4 cells at 30°C when grown on solid media containing 25 µM 5-FU. This growth defect is more evident when the cells are challenged with both 37°C and 25 µM 5-FU. As a comparison, the rrp4-G226D cells show a severe growth defect when grown on solid media containing 5-FU both at 30°C and 37°C. To further explore whether the rrp4-M68T cells exhibit other changes in cell growth, we tested for growth defects when cells are grown on media containing chemicals that disrupt different cellular pathways (Fig. 3e). Formamide alters RNA metabolism (Hoyos-Manchado et al. 2017), hydroxyurea impairs DNA synthesis (Slater 1973), and phleomycin acts as a mutagen by introducing double-strand breaks in DNA (Suzuki et al. 1970). The rrp4-M68T cells do not show any increased sensitivity when grown at 30°C on solid media containing 3% formamide, 150 mM hydroxyurea, or 5 µg/mL phleomycin (Fig. 3e). In contrast, the rrp4-G226D cells show enhanced growth defects at 30°C when grown on solid media containing 3% formamide, 150 mM hydroxyurea, or 5 µg/mL phleomycin. Taken together, these data suggest that the rrp4-M68T cells are sensitive to defects in RNA processing but do not exhibit the same extent of disrupted cellular pathways as the previously studied rrp4-G226D cells, which model a pathogenic RNA exosomopathy mutation that has severely impaired RNA exosome function in vivo (Sterrett et al. 2021).
rrp4-M68T cells have impaired RNA exosome function in processing RNA targets linked to Mtr4-RNA exosome interactions
To further assess the in vivo consequences on RNA exosome function of the rrp4-M68T variant, we measured the steady-state levels of several RNA exosome targets in rrp4-M68T cells using RT-qPCR. We assessed the steady-state levels of precursor RNAs that are targeted by the RNA exosome and are impacted by mtr4 mutant alleles, including the telomerase component RNA TLC1, which is processed by the RNA exosome in a manner dependent on TRAMP complex association, and the 3′ extended forms of U4 snRNA and snR33 snoRNA (Van Hoof et al. 2000; Houseley et al. 2006; Coy et al. 2013). In this analysis, we included both rrp4-G226D and mtr4-1 cells as comparative controls. The mtr4-1 cells have a missense mutation in MTR4 that results in accumulation of polyadenylated targets within the nucleus (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996; Weir et al. 2010). We detect increases in the steady-state level of both mature and precursor TLC1 in rrp4-M68T cells similar to that observed in rrp4-G226D cells (Fig. 4a) (Sterrett et al. 2021). Both mature and precursor TLC1 steady-state levels are significantly increased in mtr4-1 cells. Furthermore, we detect a significant increase in the steady-state level of the 3′ extended forms of the U4 snRNA and snR33 snoRNA in the rrp4-M68T cells (Fig. 4b and 4c). This increase in the levels of the extended form of these target RNAs is also observed in the rrp4-G226D cells and, to an even larger extent, the mtr4-1 cells. We also assessed steady-state levels of 5.8S rRNA precursors in rrp4-M68T and found no accumulation compared to wild-type RRP4 cells (Supplementary Fig. 3), although levels of both mature 5.8S rRNA and pre-5.8S rRNA do increase in rrp4-G226D cells which supports previous observations that the rrp4-G226D cells exhibit accumulation of 7S rRNA (Sterrett et al. 2021).
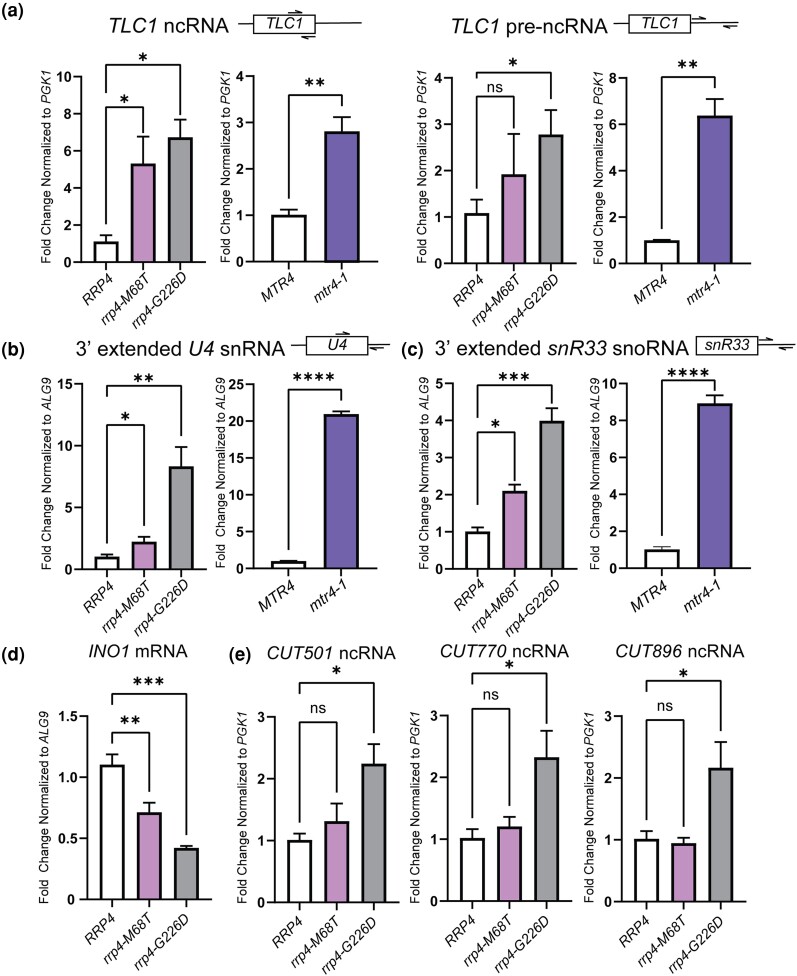
Fig. 4.
The rrp4-M68T mutant cells show elevated levels of specific RNA exosome target transcripts that depend on the Mtr4-RNA exosome interaction in vivo. The steady-state level of several RNA exosome target transcripts was assessed in rrp4-M68T cells (denoted in pink). The steady-state levels of these RNAs were also assessed in the previously described rrp4 variant rrp4-G226D as a control (denoted in gray). a) The rrp4-M68T cells show an elevated steady-state level of mature TLC1 telomerase component ncRNA relative to RRP4 cells. The steady-state level of the precursor TLC1 ncRNA in rrp4-M68T cells follows this upward trend though not statistically significant compared to RRP4 cells. This increase in both mature and precursor TLC1 is also observed in mtr4-1 (denoted in purple) compared to the wild-type control MTR4. b) The rrp4-M68T cells show an elevated steady-state level of 3′-extended pre-U4 snRNA relative to RRP4 cells. The rrp4-G226D mutant cells and mtr4-1 cells have even higher steady-state levels of this pre-U4 snRNA when compared to the RRP4 and MTR4 controls, respectively. c) The rrp4-M68T cells show an elevated steady-state level of 3′ extended snR33 snoRNA relative to RRP4 cells that is similar to the increase observed in the rrp4-G226D mutant cells and mtr4-1 cells. d) The rrp4-M68T cells show a decreased steady-state level of the mRNA transcript INO1 compared to wild-type RRP4 control cells. A decrease in this mRNA was shown previously in rrp4-G226D cells (Sterrett et al. 2021). e) The steady-state levels of non-coding, CUTs, CUT501, CUT770, and CUT896, are not significantly increased in rrp4-M68T cells compared to control as shown previously in the rrp4-G226D mutant cells (Sterrett et al. 2021). In a–e), total RNA was isolated from cells grown at 37°C and transcript levels were measured by RT-qPCR using gene-specific primers and graphed as described in Materials and Methods. Gene-specific primer sequences are summarized in Supplementary Table 2. The location of primers specific to the ncRNA transcripts are graphically represented by the cartoons above each bar graph. Within the cartoon transcript, the box represents the body of the mature transcript. Error bars represent standard error of the mean from three biological replicates. Statistical significance of the RNA levels in rrp4 variant cells relative to RRP4 cells and in the mtr4-1 cells relative to MTR4 cells is denoted by an asterisk (*P-value ≤ 0.05, **P-value ≤ 0.01, ***P-value ≤ 0.001, and ****P-value ≤ 0.0001).
We also measured the steady-state levels of select targets that are impacted within rrp4-G226D cells (Sterrett et al. 2021). We assessed the target INO1, which encodes inositol-3-phosphate synthetase (Donahue and Henry 1981; Klig and Henry 1984). INO1 mRNA has previously been identified as a transcript bound to the catalytic subunit of the RNA exosome (Delan-Forino et al. 2017) and was the most significantly decreased transcript in a previous RNA-Seq analysis of the rrp4-G226D cells (Sterrett et al. 2021). In rrp4-M68T cells, the steady-state level of INO1 is significantly decreased, similar to results for rrp4-G226D (Fig. 4d). We also assessed three CUTs that accumulate within rrp4-G226D cells (Sterrett et al. 2021). The steady-state levels of these three CUTs are not significantly increased in rrp4-M68T cells (Fig. 4e). Taken together, these data suggest that the rrp4-M68T cells have some molecular consequences resulting from the modeled multiple myeloma amino acid substitution; however, they differ from those resulting from the modeled SHRF substitution in the rrp4-G226D cells.
The Rrp4 M68T variant can associate with the RNA exosome complex
The sensitivity of the rrp4-M68T cells to drugs that impact RNA processing (Fig. 3d) and the observed accumulation of key RNA exosome target RNAs (Fig. 4) suggest that RNA exosome function may be impaired by the modeled multiple myeloma amino acid substitution. Previous studies suggest that SHRF-linked amino acid substitutions modeled in Rrp4 affect the RNA exosome function in part by disrupting complex integrity (Sterrett et al. 2021). To provide a first step toward assessing the impact of the modeled multiple myeloma amino acid substitution on the association of Rrp4 with other RNA exosome core subunits, we first assayed the protein level of Rrp4 M68T. We measured the steady-state level of the Myc-tagged Rrp4 M68T subunit when expressed as the sole copy of the Rrp4 protein in rrp4Δ cells grown at either 30°C or 37°C (Fig. 5a). Immunoblotting reveals that the steady-state level of Rrp4 M68T is comparable to wild-type Rrp4 at both temperatures tested. We next performed coimmunoprecipitations using RRP45-TAP cells that contain the endogenous, genomic RRP4 gene and express a C-terminally tandem affinity purification (TAP)-tagged Rrp45 core subunit from the endogenous RRP45 locus. We expressed Rrp4-Myc or Rrp4 M68T-Myc from plasmids in these RRP45-TAP cells. The Rrp45-TAP protein was immunoprecipitated and association of the Myc-tagged Rrp4 variants was assayed by immunoblotting (Fig. 5b). Under these conditions in which an endogenous copy of RRP4 is present, we can detect association of Rrp4 M68T-Myc with Rrp45-TAP at levels equal to that of Rrp4-Myc. As a comparative control, we also performed coimmunoprecipitations with RRP45-TAP cells expressing an exogenous Rrp4 G226D-Myc variant. Under these conditions with an endogenous copy of RRP4 present, we cannot detect association of Rrp4 G226D-Myc with the TAP-tagged core subunit, supporting previous observations (Sterrett et al. 2021). Taken together, these data suggest that Rrp4 M68T is biochemically similar to a wild-type Rrp4 subunit, and the multiple myeloma amino acid substitution likely has no impact on RNA exosome complex integrity.
Fig. 5.
The modeled multiple myeloma amino acid substitution in Rrp4 does not impact the Rrp4 protein level or association of the cap subunit with the RNA exosome complex. a) The steady-state level of the Rrp4 M68T protein variant is equal to that of wild-type Rrp4 at both 30°C and 37°C. Lysates of rrp4Δ cells expressing Myc-tagged wild-type Rrp4 or Rrp4 M68T grown at 30°C or 37°C were analyzed by immunoblotting with an anti-Myc antibody. An anti-Pgk1 antibody was used to detect 3-phosphoglycerate kinase (Pgk1) as a loading control. The mean percentage of Rrp4 M68T-Myc normalized to Rrp4-Myc from four independent experiments (n = 4) is shown. Quantitation of the immunoblot was performed as described in Materials and Methods. b) The Rrp4 M68T variant coprecipitates with the RNA exosome core subunit Rrp45 in the presence of a wild-type copy of Rrp4. Tandem affinity purification-tagged Rrp45 was immunoprecipitated from RRP45-TAP cells expressing endogenous, wild-type Rrp4, and coexpressing Myc-tagged Rrp4, Rrp4 M68T, or, as a control, Rrp4 G226D grown at 30°C and bound (top) and input (bottom) samples were analyzed by immunoblotting. As a control, immunoprecipitations were also performed from untagged RRP45 cells (no TAP control) expressing Myc-tagged Rrp4 and Rrp4 variants. The bound/input level of Rrp4-Myc was detected with anti-Myc antibody and the bound/input level of Rrp45-TAP was detected with a peroxidase–antiperoxidase (PAP) antibody. Bound Rrp45-TAP is also detected by the anti-Myc antibody as the protein A moiety of the TAP tag binds to the antibody. The input level of 3-phosphoglycerate kinase (Pgk1) was detected with an anti-Pgk1 antibody and shown as a loading control. Data shown is representative of three independent experiments (n = 3).
The rrp4-M68T mutant shows negative genetic interactions with mtr4 mutants that impact TRAMP complex association and RNA helicase unwinding
As the Rrp4 M68T variant associates with the RNA exosome complex and has a steady-state level equivalent to wild-type Rrp4, the observed sensitivity to disrupted RNA processing in rrp4-M68T cells (Fig. 3d) and accumulation of select RNA exosome target transcripts (Fig. 4) could be due to altered interaction between Rrp4 and Mtr4. As depicted in Fig. 6a, the nuclear RNA exosome cofactors Mpp6 and Rrp47 and the associated exonuclease Rrp6 aid in recruiting Mtr4 to the RNA exosome (Wasmuth et al. 2017). Rrp6 and Rrp47 form a composite site that binds to the N-terminus of Mtr4, recruiting the helicase to the RNA exosome (Schuch et al. 2014). Mtr4 forms contacts with the cap subunit Rrp4 and the cofactor Mpp6, stabilizing the helicase on the RNA exosome complex through a very conserved interface between the cap subunit and the helicase (Falk et al. 2017; Weick et al. 2018). The interaction between Mtr4 and Rrp4 provides a surface for the RNA exosome to associate with the TRAMP complex, which helps facilitates nuclear RNA surveillance and quality control of ncRNA (Lacava et al. 2005; Vaňáčová et al. 2005; Houseley and Tollervey 2008; Houseley and Tollervey 2009; Lubas et al. 2011; Stuparevic et al. 2013; Rodríguez-Galán et al. 2015; Kilchert et al. 2016; Zinder and Lima 2017; Ogami et al. 2018; Belair et al. 2019). In addition to Mtr4, the TRAMP complex is composed of a zinc-knuckle RNA binding protein, Air1 or Air2, and a non-canonical oligo(A) polymerase, Trf4 or Trf5, that oligoadenylates RNA (Belair et al. 2018). The TRAMP complex triggers degradation by adding short polyadenylated tails to the 3′ end of substrate RNA and delivering them to the RNA exosome (Houseley et al. 2006; Anderson and Wang 2009; Belair et al. 2018; Ogami et al. 2018).
Fig. 6.
The rrp4-M68T mutant cells show specific negative genetic interactions with mtr4 mutants that are predicted to impair the Trf4/5-Air1/2-Mtr4 (TRAMP) complex. a) Cartoon depicting the budding yeast nuclear RNA exosome with interacting nuclear cofactors Mpp6 (turquoise, labeled "Mpp6") and Rrp47 (dark green, labeled "Rrp47"), the exoribonuclease Rrp6 (light green, labeled "Rrp6"), and the essential RNA helicase, Mtr4 (purple) (Schuch et al. 2014; Falk et al. 2017; Schuller et al. 2018). The association of Mtr4 with the RNA exosome is facilitated by interactions between Mtr4 and Rrp6/Rrp47 (denoted by the red arrowed line) and by interactions with Mpp6 which is associated with the Rrp40 RNA exosome subunit and the Rrp4 subunit (Weir et al. 2010; Schuch et al. 2014; Wasmuth et al. 2017). The association of Mtr4 with the RNA exosome can also facilitate interaction with the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex, which triggers degradation of certain RNA targets by adding short oligo(A) tails to the 3′ end of these targets and delivering them to the RNA exosome (Houseley et al. 2006; Anderson and Wang 2009; Belair et al. 2018; Ogami et al. 2018). In addition to Mtr4, the TRAMP complex is composed of a noncanonical poly(A) polymerase, Trf4/5, and a zinc-knuckle RNA binding protein, Air1/2 (Belair et al. 2018). Central to the degradation of TRAMP-targeted RNAs by the RNA exosome is the association of Mtr4 with Trf4/5, Air1/2, and the cap subunits and nuclear cofactors of the RNA exosome complex (Falk et al. 2014; Schuch et al. 2014). b) Domain structure for S. cerevisiae Mtr4. The helicase has a low-complexity N-terminal sequence followed by the conserved helicase region. The helicase region is composed of two RecA domains and a helical domain (labeled helical bundle) that form the globular core typical of DExH family proteins. The helical bundle was originally described as the “ratchet” domain for its role in translocating nucleic acid by a Brownian ratchet (Büttner et al. 2007). In addition, Mtr4 contains an insertion domain and KOW domain that fold into a helical stalk (labeled SK insertion) (Jackson et al. 2010; Weir et al. 2010). The amino acid changes used for this experiment are labeled in red along the domain structure. c) Double mutant cells containing rrp4-M68T and specific mtr4 mutants show lethality at both 30°C and 37°C. The rrp4Δ mtr4Δ double mutant cells were serially diluted, spotted onto solid media, and grown at the indicated temperatures for 3 days. The mtr4 mutant plasmids included in this experiment are as follows; mtr4-1—a temperature-sensitive mutant that contains a missense mutation resulting in the amino acid substitution Cys942Tyr, which causes accumulation of poly(A)+ RNA in the nucleus at 37°C (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996); mtr4-F7A-F10A—an mtr4 allele that impairs the interaction with Rrp6/Rrp47 (Schuch et al. 2014); mtr4-R349E-N352E—a mutation that impairs the association of Mtr4 with the poly(A) RNA polymerase Trf4 with the Mtr4 helicase (Falk et al. 2014); mtr4R-1030A and mtr4-E1033W—two mutations within the helical bundle that differentially impact nucleic acid unwinding by Mtr4 (Taylor et al. 2014). mtr4-1 mutant cells expressing RRP4, rrp4-M68T, or rrp4-G226D show lethality at 37°C presumably due to the known temperature-sensitive nature of the mtr4-1 allele (Liang et al. 1996). Growth of double mutant cells containing rrp4-M68T or rrp4-G226D is shown. d) Summary of rrp4 mtr4 mutant cell growth. Triplicate solid media assays were performed on double mutant cells containing rrp4-M68T or rrp4-G226D and the series of mtr4 variants. Cell growth at both 30°C and 37°C was semiquantified on a scale of zero (no growth; black) to five (comparable to RRP4 wild-type growth; white). Growth scale of the double mutant cells is represented through the color gradient on the two heatmaps. All double mutant cells were generated as described in Materials and Methods. Images shown are from a singular solid media growth assay with all samples plated on the same Leu− media plate. Data are representative of three independent experiments (n = 3).
To assess genetic interactions between Mtr4 and the RNA exosome in S. cerevisiae modeling the multiple myeloma patient mutation, we performed an analysis of a series of five mtr4 mutant alleles that introduce amino acid substitutions in Mtr4 as summarized in Fig. 6b. We also included the rrp4 mutant variant, rrp4-G226D, for comparison as this rrp4 variant has a negative interaction with the mtr4-F7A-F10A mutant allele (Sterrett et al. 2021). Included within our five mtr4 alleles is the mtr4-1 allele, a known temperature sensitive mutant (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996; Weir et al. 2010). These genetic data are shown both as representative solid growth assays (Fig. 6c) and as a heatmap (Fig. 6d), which summarizes data from three independent replicates for all these genetic experiments. As predicted, RRP4 mtr4-1 cells have a severe growth defect at 37°C that is shared by both double mutant rrp4-M68T mtr4-1 and rrp4-G226D mtr4-1.
Two of the five mtr4 mutant alleles, mtr4-F7A-F10A and mtr4-R349E-N352E, impair protein–protein interactions of Mtr4 in S. cerevisiae. The Mtr4 F7A F10A variant disrupts Mtr4 interactions with Rrp6/Rrp47 by introducing two amino acid substitutions, F7A and F10A, into the N-terminus of Mtr4 (Fig. 6b) (Schuch et al. 2014). The mtr4-R349E-N352E mutant allele impairs the association of Mtr4 with the poly(A) RNA polymerase Trf4 within the TRAMP complex and thus disrupts the recruitment of the TRAMP complex to the RNA exosome (Falk et al. 2014). The rrp4-M68T mtr4-F7A-F10A double mutant cells grow similarly to rrp4-M68T cells at 30°C; however, at 37°C, the double mutant cells show a mild growth defect in comparison with the single mutant rrp4-M68T and the RRP4 mtr4-F7A-F10A cells (Fig. 6d). As shown previously, the rrp4-G226D mtr4-F7A-F10A cells show a severe growth defect at 37°C compared to rrp4-G226D cells (Sterrett et al. 2021). The rrp4 M68T mtr4-R349E-N352E double mutant cells show severe growth defects at 30°C and 37°C compared to the single mutant rrp4-M68T cells or the RRP4 mtr4-R349E-N352E control cells. In contrast, the rrp4 G226D mtr4-R349E-N352E double mutant cells show no difference in growth at 30°C compared to the RRP4 mtr4-R349E-N352E control cells and improved growth compared to the single mutant rrp4-G226D cells at 37°C (Fig. 6c and 6d).
The final two mtr4 mutant alleles that we tested for genetic interaction with the rrp4-M68T variant impact nucleic acid unwinding by Mtr4 (Taylor et al. 2014). Studies of an RNA-bound Mtr4 structure demonstrate that residues of R1030 and E1033 mediate key nucleic acid base interactions with the helicase helical bundle (Weir et al. 2010; Taylor et al. 2014). Mutagenesis of these residues in S. cerevisiae, generating the mutant alleles mtr4-R1030A and mtr4-E1033A, reveals that these residues play important but distinct roles in helicase activity (Taylor et al. 2014). The rrp4-M68T mtr4-R1030A double mutant cells are not viable at either temperature tested. In contrast, the rrp4-M68T mtr4-E1033A double mutant cells are viable and grow similar to the RRP4 mtr4-E1033A control cells at both 30°C and 37°C. The growth defect of rrp4-G226D at 37°C is too severe to assess genetic interactions with either the mtr4-R1030A or the mtr4-E1033A mutation under these growth conditions. However, in contrast to the lethality observed for the rrp4-M68T mtr4-R1030A double mutant, the rrp4-G226D mtr4-R1030A and rrp4-G226D mtr4-E1033A double mutant cells have comparable growth to the single mutant rrp4-G226D cells as well as the RRP4 mtr4-R1030A and RRP4 mtr4-E1033A control cells at 30°C. The rrp4-M68T double mutants that show synthetical lethality are viable when rescued by expression of a wild-type RRP4 plasmid (Supplementary Fig. 4), demonstrating that the growth defects and lethality observed are due to negative genetic interactions between the rrp4 and mtr4 mutants. Taken together, these data suggest that the rrp4-M68T cells have negative genetic interactions with specific mtr4 mutant alleles, distinct from those previously described for the rrp4-G226D mutant model.
The rrp4-M68T mutant shows negative genetic interactions with mpp6Δ
As depicted in Fig. 6a, the nuclear RNA exosome cofactors Mpp6 and Rrp47 and the exoribonuclease Rrp6 help to recruit and stabilize the interaction with Mtr4. The exosome cofactor Rrp47 interacts with and stabilizes the exoribonuclease Rrp6 and the cofactor Mpp6 interacts with the nuclear RNA exosome through direct contacts with the cap subunit Rrp40 (Schuch et al. 2014; Wasmuth et al. 2014; Wasmuth et al. 2017). To further evaluate the impact that the modeled multiple myeloma amino acid substitution may have on the RNA exosome-Mtr4 interaction in vivo, we tested whether the rrp4-M68T variant exhibits genetic interactions with mpp6 or rrp47 mutants by deleting these non-essential, nuclear exosome cofactor genes MPP6 and RRP47 in combination with rrp4-M68T. For comparison, we included the rrp4-G226D variant as these cells have known negative genetic interactions with these mutants (Sterrett et al. 2021). We examined the growth of these double mutants relative to single mutants (rrp4-M68T or rrp4-G226D) and the control mutant cells (RRP4 mpp6Δ or RRP4 rrp47Δ) in solid and liquid media growth assays (Fig. 7). In the solid media growth assays, the rrp4-M68T mpp6Δ double mutant cells show growth very similar to the rrp4-M68T cells at both 30°C and 37°C after both 1 and 2 days of growth (Fig. 7b). The rrp4-M68T rrp47Δ cells show a severe growth defect at 37°C compared to the single mutant rrp4-M68T; however, this impaired growth is comparable to that of the RRP4 rrp47Δ cells, which has been previously reported for the single mutant rrp47Δ (Mitchell et al. 2003) (Fig. 6c). In contrast, the rrp4-G226D mpp6Δ and rrp4-G226D rrp47Δ double mutant cells show a severe growth defect at both temperatures compared to the single mutant rrp4-G226D cells as described previously (Sterrett et al. 2021).
Fig. 7.
The rrp4-M68T mpp6Δ double mutant cells exhibit impaired growth that is exacerbated on drugs that impact RNA processing. a) Cartoon schematic of the budding yeast nuclear RNA exosome in complex with nuclear cofactors Mpp6 (turquoise, labeled "Mpp6") and Rrp6/47 (light green/dark green, labeled "Rrp6" and "Rrp47") (Schuller et al. 2018). Serial dilution growth assays of double mutant (B) rrp4-M68T mpp6Δ or (C) rrp4-M68T rrp47Δ cells at 30°C and 37°C. The double mutant cells (rrp4Δ with mpp6Δ or rrp47Δ) containing control RRP4 or rrp4 variants rrp4-M68T and rrp4-G226D plasmids were serially diluted, spotted onto selective solid media, and grown at the indicated temperatures for two days. The double mutant cells rrp4-G226D mpp6Δ and rrp4-G226D rrp47Δ were included as a comparative control and show growth defects as described previously (Sterrett et al. 2021). Data shown are representative of three independent assays (n = 3). d) and e) Double mutant cells containing rrp4-G226D and mpp6Δ exhibit a statistically significant increase in doubling time in liquid culture. Double mutant cells (rrp4Δ mpp6Δ or rrp4Δ rrp47Δ) containing control RRP4 or rrp4-M68T plasmids were diluted in selective media and grown at 37°C with optical density measurements used to assess cell density over time. Data shown is collected from four independent samples (n = 4). e) Doubling time for each sample was quantified and normalized to the growth rate of control RRP4 cells. All calculations were performed as described in Materials and Methods. Full liquid growth curves of both rrp4-M68T mpp6Δ and rrp4-M68T rrp47Δ mutant cells are shown in Supplementary Fig. 5. f) Double mutant cells rrp4-M68T mpp6Δ exhibit impaired growth on solid media containing drugs impacting RNA processing. The rrp4Δ mpp6Δ cells expressing RRP4, rrp4-M68T, or rrp4-G226D were serially diluted, spotted onto solid YEPD media containing 3% formamide or selective media containing 25 µM fluorouracil (5-FU) and grown at 30°C for three days. The rrp4-M68T mpp6Δ cells show impaired growth when compared to RRP4 mpp6Δ cells. The rrp4-G226D mpp6Δ cells show exacerbated growth defects on 3% formamide and 25 µM 5-FU at 30°C. Data shown are representative of three independent assays (n = 3).
While the solid media growth assay suggests comparable growth between the controls and the rrp4-M68T double mutant cells, the liquid media growth assay reveals a modest growth defect of the rrp4-M68T mpp6Δ at 37°C compared to both the rrp4-M68T and control RRP4 mpp6Δ cells (Fig. 7d), with the doubling time significantly longer than that of wild-type RRP4 cells (Fig. 7e). The liquid growth assay also shows doubling times for rrp4-M68T rrp47Δ and RRP4 rrp47Δ double mutants are nearly twice that of wild-type RRP4 cells but do not differ significantly when compared to each other (Fig. 7d and 7e). The observed growth defect of the rrp4-M68T mpp6Δ double mutant in liquid culture is revealed in a solid media growth assay when the cells are challenged with formamide or 5-FU (Fig. 7f). The distinct growth defect of the rrp4-G226D mpp6Δ double mutant is also exacerbated by growth on these chemicals. Taken together, these data suggest a negative genetic interaction between the rrp4 variants and mpp6 mutants, with both double mutants having exacerbated defects when challenged with drugs that impact RNA metabolism.
The Rrp4 M68T variant shows decreased association with the essential helicase Mtr4
Given the negative genetic interactions of rrp4-M68T with both mpp6 and mtr4 mutants and the physical interactions between both EXOSC2 Met40 and Rrp4 Met68 and the RNA helicase MTR4/Mtr4 in the RNA exosome structures (Schuller et al. 2018; Weick et al. 2018), we predicted that the physical interaction between the RNA exosome and Mtr4 would be affected in rrp4-M68T cells. Previous studies investigated the physical interaction between Rrp4 G226D and Mtr4 using a coimmunoprecipitation assay and the data suggested that there is decreased association between the RNA exosome and the helicase in rrp4-G226D cells (Sterrett et al. 2021). We employed a similar approach to investigate whether the physical interaction between Mtr4 and the RNA exosome is impacted by the Rrp4 M68T variant. We performed a coimmunoprecipitation with cells expressing Rrp4-Myc or Rrp4 M68T-Myc as the sole copy of Rrp4 and coexpressing Mtr4-FLAG (Fig. 8). The Rrp4-Myc proteins were immunoprecipitated and association with Mtr4-FLAG was assayed by immunoblotting. As shown in Fig. 8, there is a significant decrease in the amount of Mtr4-FLAG that coimmunoprecipitates with Rrp4 M68T-Myc as compared to Rrp4-Myc (Fig. 8a-b). The amount of Rrp4 M68T-Myc and Rrp4-Myc in both the input and immunoprecipitation is comparable (Fig. 8c), showing that the difference in detected Mtr4-FLAG is not due to decreased protein levels or inefficient immunoprecipitation of Rrp4 M68T-Myc (Fig. 8c). These data, therefore, suggest that Mtr4 association with the Rrp4 cap subunit is significantly disrupted by the Rrp4 M68T amino acid substitution. Combined with the genetic data (Fig. 6 and Fig. 7), the structural modeling (Fig. 2), and the increased steady-state level of RNA exosome target RNAs (Fig. 4), these results strongly suggest that there is destabilization of the interaction between Mtr4 and the RNA exosome complex in rrp4-M68T cells that impacts the function of the molecular machine.
Fig. 8.
Rrp4 M68T shows decreased association with Mtr4 compared to wild-type Rrp4. a) The rrp4Δ cells coexpressing Rrp4-Myc variants and FLAG-tagged Mtr4 were grown at 30°C. Myc-tagged Rrp4 or Rrp4 M68T protein was immunoprecipitated from cleared lysate using anti-Myc beads and bound Mtr4-FLAG protein was detected by immunoblotting. The bound/input level of Mtr4-FLAG was detected with an anti-FLAG antibody and the bound/input level of Rrp4-Myc was detected with an anti-Myc antibody. The 3-phosphoglycerate kinase (Pgk1) serves as a loading control. b) Quantitation of the percentage of bound to input Mtr4-FLAG coimmunoprecipitated with Rrp4-Myc or Rrp4 M68T-Myc normalized to Pgk1. The graph shows the mean percentage of bound Mtr4-FLAG normalized to unbound input. Error bars represent standard error of the mean. c) Quantitation of percentage of bound to input Rrp4-Myc or Rrp4 M68T-Myc immunoprecipitated normalized to Pgk1. Error bars represent standard error of the mean. Statistical significance is denoted (*P-value ≤ 0.05; n.s. P-value ≥ 0.05). Data shown here collected were from two independent experiments (n = 2). The coimmunoprecipitations were performed and quantitated as described in Materials and Methods.
Discussion
In this study, we modeled and analyzed a multiple myeloma patient EXOSC2 mutation in the S. cerevisiae homolog RRP4. We generated rrp4-M68T mutant cells expressing the variant Rrp4 M68T, which corresponds to the EXOSC2 M40T variant. Analysis of these rrp4-M68T cells reveals that this amino acid substitution affects RNA exosome function. While our biochemical assays show that the Rrp4 M68T variant can associate with the RNA exosome complex and function as the sole copy of the essential Rrp4 RNA exosome cap subunit, rrp4-M68T cells do show growth defects when grown in media containing drugs that impact RNA processing. The rrp4-M68T cells also show accumulation of known RNA exosome targets. These defects in RNA exosome function could result from an impaired interaction between the complex and the essential RNA helicase Mtr4 as predicted by structural modeling. Our genetic analyses support this model as rrp4-M68T cells show negative genetic interactions with both mpp6 and mtr4 mutants. Furthermore, we demonstrate through a coimmunoprecipitation assay that the M68T substitution in Rrp4 decreases association with the Mtr4 helicase. These data suggest that the introduction of the multiple myeloma-associated amino acid change could impact the binding interface between EXOSC2 and MTR4, potentially impairing the function of the essential RNA exosome in vivo for a subset of Mtr4-dependent targets.
Structural studies reveal the evolutionary conservation of the interaction between the RNA exosome and Mtr4 (Supplementary Fig. 2), with the helicase cofactor interacting with the complex through multiple points of contact, including a direct interface with EXOSC2/Rrp4 and indirect stabilizing interactions with the cofactors Mpp6, Rrp47, and the associated exonuclease Rrp6 (Falk et al. 2017; Weick et al. 2018). This robust interaction between the complex and the essential helicase likely explains why the rrp4-M68T cells show no functional consequences unless challenged through introduction of drugs impacting RNA processing or loss of other stabilizing cofactors, such as in rrp4-M68T mpp6Δ double mutant cells. While this model would also predict a negative genetic interaction between rrp47Δ and rrp4-M68T, the rrp4-M68T rrp47Δ cells show a growth defect at 37°C that is indistinguishable from that of RRP4 rrp47Δ cells. This growth defect in rrp4-M68T rrp47Δ and RRP4 rrp47Δ cells is likely due to the loss of Rrp6 association with the RNA exosome complex given the stabilizing role that Rrp47 plays for Rrp6 (Mitchell et al. 2003; Wasmuth et al. 2014). The growth defects resulting from destabilization of Mtr4 in rrp4-M68T rrp47Δ cells are likely masked by the larger consequence of disassociating Rrp6 from the complex. We do detect a slight growth defect in rrp4-M68T cells expressing an mtr4 variant that disrupts the stabilizing interactions between Rrp6, Rrp47, and Mtr4 (mtr4-F7A-F10A), pointing to the importance of the Rrp4-Mtr4 interface. We do, however, observe significant molecular consequences in the rrp4-M68T cells. We detect accumulation of several documented RNA exosome target transcripts, particularly those linked to RNA exosome–Mtr4 association (Van Hoof et al. 2000; Houseley et al. 2006). Using a biochemical assay, we also observe a significant decrease in interaction between Mtr4 and Rrp4 M68T as compared to wild-type Rrp4. This decreased association further suggests that the modeled multiple myeloma mutation destabilizes the interaction between the RNA exosome and the essential RNA helicase. Taken together, these data suggest that while the consequences resulting in vivo from the Rrp4 M68T variant are subtle at the macro scale, they are impactful molecularly for a specific set of target RNAs and for the biochemical interaction between the RNA exosome and Mtr4.
The interaction between the RNA exosome and Mtr4 could also be critical for other interactions, particularly those involving the TRAMP (Trf4/5-Air1/2-Mtr4 polyadenylation) complex. Our genetic analyses reveal a negative genetic interaction between rrp4-M68T and mtr4-R349E-N352E. The Mtr4 R349E N352E variant impairs Mtr4-Trf4 binding and impacts TRAMP complex assembly in vivo (Falk et al. 2014). The rrp4-M68T cells that express Mtr4 R349E N352E as the sole copy of the helicase grow very poorly at both 30°C and 37°C as compared to control RRP4 mtr4-R349E-N352E cells, suggesting that TRAMP complex assembly and association with the RNA exosome may also be impacted by Rrp4 M68T. Intriguingly, we also detect synthetic lethality for the rrp4-M68T mtr4-R1030A double mutant. This lethality is specific to rrp4-M68T mtr4-R1030A cells as the rrp4-M68T cells expressing the other helicase mutant, mtr4-E1033W, show growth similar to the control (RRP4 mtr4-E1033W). Both Mtr4 R1030A and Mtr4 E1033W decrease helicase unwinding capability (Taylor et al. 2014). However, the Mtr4 R1030A variant is also implicated in disrupting target discrimination by the TRAMP complex, potentially by disrupting preferential polyadenylation by Trf4 (Taylor et al. 2014). Therefore, the negative genetic interaction observed for rrp4-M68T rrp4-R1030A cells further suggests that TRAMP function is impacted in rrp4-M68T cells. Taken together with our structural modeling data, we hypothesize that a stabilized interaction between the RNA exosome and Mtr4 is necessary for TRAMP association and the slightest perturbation, even a subtle destabilization at one contact point with the helicase, could disrupt this vital interaction between TRAMP and the complex. More biochemical studies could be performed to explore how changes within the EXOSC2-MTR4/Rrp4-Mtr4 interface impact the interaction with the TRAMP complex.
Our studies also show that rrp4-M68T mutant cells have distinct genetic interactions as compared to the rrp4-G226D cells. The rrp4-G226D mtr4-R349E-N352E double mutant cells surprisingly show improved growth at 37°C compared to either single mutant. Even more surprising is the synthetic lethality in cells expressing rrp4-G226D and either mtr4 helicase mutant (mtr4-R1030A and mtr4-E1033W). These genetic interactions could suggest that the modeled SHRF amino acid substitution (Rrp4 G226D) has distinct in vivo consequences compared to the modeled multiple myeloma-associated substitution Rrp4 M68T. The Rrp4 G226D variant has decreased association with Mtr4 and the rrp4-G226D cells show transcriptomic differences from wild-type cells consistent with disrupted RNA exosome-Mtr4 interactions (Sterrett et al. 2021). Similarly, we observe decreased association between the Rrp4 M68T variant and Mtr4 and some RNA exosome target transcripts accumulate in rrp4-M68T cells that also accumulate in rrp4-G226D cells. However, notably, we do not detect any changes in select CUTs or 5.8S rRNA precursors in rrp4-M68T cells. We do, intriguingly, observe a significant decrease in the steady-state level of INO1 mRNA in rrp4-M68T cells that is shared in the rrp4-G226D cells (Sterrett et al. 2021). Previous work characterizing the rrp4-G226D mutation suggested that the significant change in INO1 mRNA levels could reflect defects in the cytoplasmic roles of the RNA exosome (Sterrett et al. 2021), though the exact molecular mechanism remains unknown. A comparison of the results obtained for rrp4-M68T and rrp4-G226D suggests that there may be some distinct defects in RNA exosome function in each of these mutants though they may also have some overlapping consequences in vivo, in part due to altered association with Mtr4. The difference in molecular consequences between the two rrp4 variants could be attributed to the impact on RNA exosome complex integrity observed in rrp4-G226D cells, which was not observed in rrp4-M68T cells (Fig. 5b) (Sterrett et al. 2021).
The difference in severity of functional and molecular consequences that we observe for the rrp4-M68T and rrp4-G226D mutant models may partially explain the differences in disease pathology between SHRF patients with the mutation EXOSC2 G198D and the multiple myeloma patient with the mutation EXOSC2 M40T. The EXOSC2 G198D mutation was identified in SHRF patients through whole exome sequencing and classified as causing a novel Mendelian syndrome (Di Donato et al. 2016). In contrast, the EXOCS2 M40T mutation is a spontaneous, somatic mutation that likely cooccurred with a chromosome 9 duplication. Additionally, the patient with this EXOSC2 M40T mutation has several chromosomal aberrations that are a hallmark of multiple myeloma, suggesting that these EXOSC2 mutations could be passenger mutations rather than a pathogenic driver of the multiple myeloma. Upon further analysis of the non-coding mutations in the patient harboring EXOSC2 M40T, we found a second mutation present in intron 1 of EXOSC2 in this patient (Supplementary Fig. 6). This mutation (EXOSC2 SNV chr9:130,693,915 T > G) is predicted to alter the splice donor site and likely to result in a misprocessed mRNA or truncated protein. Through RNA-Seq data available in CoMMpass for this patient, we determined that the EXOSC2 M40T missense mutation and the splice donor mutation are expressed from the same allele. Interestingly, we calculate the allelic frequency of these two EXOSC2 mutations to be very similar (0.2266 vs. 0.2191). This suggests that EXOSC2 M40T and the EXOSC2 splice donor mutation either cooccurred or that the splice donor mutation was selected for in response to the EXOSC2 M40T missense mutation, which could negatively affect cell growth and/or survival. As EXOCS2 is an essential gene in 1,076/1,086 cancer cell lines in the Cancer Dependencies Map project (depmap.org) including all 19 myeloma cell lines in the dataset, a future approach would be to engineer the EXOSC2 M40T mutation into myeloma cell lines to determine the effects on RNA exosome function as well as myeloma cell growth and survival.
As the rrp4-M68T cells show defects in RNA exosome function, likely through altered of interactions with the RNA helicase Mtr4 and the associated TRAMP complex, this EXOSC2 M40T substitution could be detrimental to the function of the human RNA exosome. Altering key cofactor interactions with the RNA exosome could impact the processing and degradation of target RNA transcripts such as small ncRNA species that have key regulatory roles in various cellular processes. Furthermore, the interaction between the RNA exosome and MTR4 has been suggested to resolve secondary DNA structures associated with strand asymmetric DNA mutagenesis that can lead to genome instability and chromosomal translocations particularly in plasma B cells (Lim et al. 2017). The high level of evolutionary conservation within the N-terminus of EXOSC2 that interacts with MTR4 (Fig. 1c and Supplementary Fig. 2) suggests that there could be evolutionary pressure to maintain the integrity of certain sequences within EXOSC2 that specifically interact with key cofactors. Taking a genetic approach to assess different EXOSC missense mutations associated with human diseases can help unravel different consequences in specific interactions of the essential RNA exosome complex.
Utilizing the yeast genetic model system, we have characterized an EXOSC2 mutation found in a multiple myeloma patient. However, this mutation was one of several mutations identified in genes encoding structural subunits of the RNA exosome in the CoMMpass study (Supplementary Fig. 1). The frequency of multiple myeloma mutations identified in the DIS3 catalytic exosome gene suggests that there is an important link between multiple myeloma and the RNA exosome. By modeling identified EXOSC mutations in the budding yeast system, we can examine whether these mutations impair the function of the essential RNA exosome and provide a deeper understanding of the role that this conserved complex may have in cancer pathologies. While it is unlikely that the identified EXOSC mutations drive the multiple myeloma disease, our study here clearly shows that these mutations have in vivo consequences for the conserved and essential RNA exosome-Mtr4 interaction. In addition, by studying other models of EXOSC disease-linked mutations, such as those identified in RNA exosomopathy patients, we can provide insight into the biological pathways that are altered in these different disorders. As more pathogenic mutations are uncovered in EXOSC genes via accelerated genomic screening of patients, generating in vivo models to explore the consequences of these changes can help to define the most critical interactions of the complex with various cofactors and thus expand our understanding of the biological functions of this essential RNA processing and degradation complex.
Supplementary Material
Acknowledgements
We thank members of the Corbett lab for critical discussions and input. We thank Dr. Benjamin Barwick for his contributions in assisting with the analysis of the CoMMpass dataset. We also thank Dr. Graeme Conn and Dr. Ambro van Hoof for their intellectual contributions.
Contributor Information
Maria C Sterrett, Department of Biology, Emory University, Atlanta, GA 30322, USA; Biochemistry, Cell, and Developmental Biology Graduate Program, Emory University, Atlanta, GA 30322, USA.
Daniela Farchi, Department of Biology, Emory University, Atlanta, GA 30322, USA.
Sarah E Strassler, Biochemistry, Cell, and Developmental Biology Graduate Program, Emory University, Atlanta, GA 30322, USA; Department of Biochemistry, Emory University, Atlanta, GA, 30322, USA.
Lawrence H Boise, Department of Hematology and Medical Oncology, School of Medicine, Emory University, Atlanta, GA 30322, USA; Winship Cancer Institute, Emory University, Atlanta, GA 30322, USA.
Milo B Fasken, Department of Biology, Emory University, Atlanta, GA 30322, USA.
Anita H Corbett, Department of Biology, Emory University, Atlanta, GA 30322, USA.
Data availability
Strains and plasmids summarized in Supplementary Table 1 and Supplementary Table 2 are available upon request. Genomic data from CoMMpass are available at dbGaP with the accession number phs000748.v7.p4. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.
Funding
This work was supported by National Institutes of Health (NIH) R01 grants (GM058728) and the NIH-funded Emory Initiative for Maximizing Student Development (R25 GM099644) to A.H.C. and National Cancer Institute (NCI) R21 grant (CA273773) to L.H.B. M.C.S. was supported by a National Institute of General Medical Sciences (NIGMS) F31 grant (GM134649-01). S.E.S. was supported by the National Science Foundation (NSF) Graduate Research Fellowship (GRFP 1937971). Lastly, we would also like to thank the Genetics Society of America and the Saccharomyces Genomic Database (SGD) (Cherry et al. 2012) for providing community resources and support for scientific discovery.
Literature cited
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Alexander DD, Mink PJ, Adami HO, Cole P, Mandel JS, Oken MM, Trichopoulos D. Multiple myeloma: a review of the epidemiologic literature. Int J Cancer. 2007;120(S12):40–61. doi: 10.1002/ijc.22718. [DOI] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, et al. . Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999;18(19):5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44(1):16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. Consurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. Consurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server):W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwick BG, Neri P, Bahlis NJ, Nooka AK, Dhodapkar MV, Jaye DL, Hofmeister CC, Kaufman JL, Gupta VA, Auclair D, et al. . Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat Commun. 2019;10(1):1911. doi: 10.1038/s41467-019-09555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belair C, Sim S, Kim KY, Tanaka Y, Park IH, Wolin SL. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J Cell Biol. 2019;218(8):2564–2582. doi: 10.1083/jcb.201811148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belair C, Sim S, Wolin SL. Noncoding RNA surveillance: the ends justify the means. Chem Rev. 2018;118(8):4422–4447. doi: 10.1021/acs.chemrev.7b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsagel PL, Nardini E, Brents L, Chesi M, Kuehl WM. Igh translocations in multiple myeloma: a nearly universal event that rarely involves c-myc. Curr Top Microbiol Immunol. 1997;224:283–287. doi: 10.1007/978-3-642-60801-8_30. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, Mueller JS, Pyle A, Munkley J, Dor T, Quartararo J, Ferrero I, Karcagi V, Giunta M, Polvikoski T, et al. . EXOSC8 Mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat Commun. 2014;5(1):4287. doi: 10.1038/ncomms5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EM, Ashby C, Tytarenko RG, Deshpande S, Wang H, Wang Y, Rosenthal A, Sawyer J, Tian E, Flynt E, et al. . BRAF and DIS3 mutations associate with adverse outcome in a long-term follow-up of patients with multiple myeloma. Clin Cancer Res. 2020;26(10):2422–2432. doi: 10.1158/1078-0432.CCR-19-1507. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273(21):13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Burns D, Donkervoort D, Bharucha-Goebel D, Giunta M, Munro B, Scavina M, Foley R, Müller JS, Bönnemann CG, Horvath R. A recessive mutation in EXOSC9 causes abnormal RNA metabolism resulting in a novel form of cerebellar hypoplasia/atrophy with early motor neuronopathy. Neuromuscul Disord. 2017;27:S38. doi: 10.1016/S0960-8966(17)30331-0. [DOI] [Google Scholar]
- Burns DT, Donkervoort S, Muller JS, Knierim E, Bharucha-Goebel D, Faqeih EA, Bell SK, AlFaifi AY, Monies D, Millan F, et al. . Variants in EXOSC9 disrupt the RNA exosome and result in cerebellar atrophy with spinal motor neuronopathy. Am J Hum Genet. 2018;102(5):858–873. doi: 10.1016/j.ajhg.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. Consurf: using evolutionary data to raise testable hypotheses about protein function. Israel J Chem. 2013;53(3–4):199–206. doi: 10.1002/ijch.201200096. [DOI] [Google Scholar]
- Cervelli T, Galli A. Yeast as a tool to understand the significance of human disease-associated gene variants. Genes (Basel). 2021;12(9):1303. doi: 10.3390/genes12091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet J-P, Ahmann GJ, Adli M, et al. . Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. . Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(D1):D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy S, Volanakis A, Shah S, Vasiljeva L. The Sm complex is required for the processing of non-coding RNAs by the exosome. PloS one. 2013;8(6):e65606–e65606. doi: 10.1371/journal.pone.0065606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da B, Dawson D, Stearns T, Methods in yeast genetics: a cold spring harbor laboratory course manual2000.
- de Amorim J, Slavotinek A, Fasken MB, Corbett AH, Morton DJ. Modeling pathogenic variants in the RNA exosome. RNA & Dis. 2020;7:e1166. PMCID: PMC8528344 [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. Embo j. 1998;17(4):1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delan-Forino C, Schneider C, Tollervey D. Transcriptome-wide analysis of alternative routes for RNA substrates into the exosome complex. PLOS Genet. 2017;13(3):e1006699. doi: 10.1371/journal.pgen.1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Donato N, Neuhann T, Kahlert A-K, Klink B, Hackmann K, Neuhann I, Novotna B, Schallner J, Krause C, Glass IA, et al. . Mutations in EXOSC2 are associated with a novel syndrome characterised by retinitis pigmentosa, progressive hearing loss, premature ageing, short stature, mild intellectual disability and distinctive gestalt. J Med Genet. 2016;53(6):419–425. doi: 10.1136/jmedgenet-2015-103511. [DOI] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981;256(13):7077–7085. doi: 10.1016/S0021-9258(19)69102-7. [DOI] [PubMed] [Google Scholar]
- Eggens VRC, Barth PG, Niermeijer J-MF, Berg JN, Darin N, Dixit A, Fluss J, Foulds N, Fowler D, Hortobágyi T, et al. . EXOSC3 Mutations in pontocerebellar hypoplasia type 1: novel mutations and genotype-phenotype correlations. Orphanet J Rare Dis. 2014;9(1):23. doi: 10.1186/1750-1172-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Bonneau F, Ebert J, Kogel A, Conti E. Mpp6 incorporation in the nuclear exosome contributes to RNA channeling through the Mtr4 helicase. Cell Rep. 2017;20(10):2279–2286. doi: 10.1016/j.celrep.2017.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Weir JR, Hentschel J, Reichelt P, Bonneau F, Conti E. The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol Cell. 2014;55(6):856–867. doi: 10.1016/j.molcel.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Fasken MB, Losh JS, Leung SW, Brutus S, Avin B, Vaught JC, Potter-Birriel J, Craig T, Conn GL, Mills-Lujan K, et al. . Insight into the RNA exosome complex through modeling pontocerebellar hypoplasia type 1b disease mutations in yeast. Genetics. 2017;205(1):221–237. doi: 10.1534/genetics.116.195917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Morton DJ, Kuiper EG, Jones SK, Leung SW, Corbett AH. The RNA exosome and human disease. Methods Mol Biol. 2020;2062:3–33. doi: 10.1007/978-1-4939-9822-7_1. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci U S A. 2004;101(3):793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie A, Gabunilas J, Jen JC, Chanfreau GF. Mutations of EXOSC3/Rrp40p associated with neurological diseases impact ribosomal RNA processing functions of the exosome in S. cerevisiae. RNA. 2017;23(4):466–472. doi: 10.1261/rna.060004.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J, Scott Butler J. Evidence for distinct DNA- and RNA-based mechanisms of 5-fluorouracil cytotoxicity in Saccharomyces cerevisiae. Yeast. 2007;24(10):861–870. doi: 10.1002/yea.1516. [DOI] [PubMed] [Google Scholar]
- Hou D, Ruiz M, Andrulis ED. The ribonuclease Dis3 is an essential regulator of the developmental transcriptome. Bmc Genomics. 2012;13(1):359. doi: 10.1186/1471-2164-13-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7(7):529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779(4):239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136(4):763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Hoyos-Manchado R, Reyes-Martín F, Rallis C, Gamero-Estévez E, Rodríguez-Gómez P, Quintero-Blanco J, Bähler J, Jiménez J, Tallada VA. RNA metabolism is the primary target of formamide in vivo. Sci Rep. 2017;7(1):15895. doi: 10.1038/s41598-017-16291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29(13):2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, et al. . Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126(3):649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Schneiter R, Hitomi M, Tartakoff AM. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6(9):1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat Rev Mol Cell Biol. 2016;17(4):227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- Klig LS, Henry SA. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc Natl Acad Sci U S A. 1984;81(12):3816–3820. doi: 10.1073/pnas.81.12.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyade nylation complex. Cell. 2005;121(5):713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Laubach J, Richardson P, Anderson K. Multiple myeloma. Annu Rev Med. 2011;62(1):249–264. doi: 10.1146/annurev-med-070209-175325. [DOI] [PubMed] [Google Scholar]
- Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16(9):5139–5146. doi: 10.1128/MCB.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SJ, Boyle PJ, Chinen M, Dale RK, Lei EP. Genome-wide localization of exosome components to active promoters and chromatin insulators in Drosophila. Nucleic Acids Res. 2013;41(5):2963–2980. doi: 10.1093/nar/gkt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Giri PK, Kazadi D, Laffleur B, Zhang W, Grinstein V, Pefanis E, Brown LM, Ladewig E, Martin O, et al. . Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell. 2017;169(3):523–537 e515. doi: 10.1016/j.cell.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. . Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8(5):470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losh J. Identifying Subunit Organization and Function of the Nuclear RNA Exosome Machinery, Houston (TX): University of Texas, UT GSBS Dissertations and Theses2018. [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43(4):624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgaertner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495(7439):70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol. 2008;28(17):5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol. 2003;23(19):6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′->5′ exoribonucleases. Cell. 1997;91(4):457–466. doi: 10.1016/S0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10(4):502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Morton DJ, Jalloh B, Kim L, Kremsky I, Nair RJ, Nguyen KB, Rounds JC, Sterrett MC, Brown B, Le T, et al. . A drosophila model of pontocerebellar hypoplasia reveals a critical role for the RNA exosome in neurons. PLoS Genet. 2020;16(7):e1008901. doi: 10.1371/journal.pgen.1008901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007;8(1):63–69. doi: 10.1038/sj.embor.7400856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Chen Y, Manley JL. RNA Surveillance by the nuclear RNA exosome: mechanisms and significance. Noncoding RNA. 2018;4(1):8. doi: 10.3390/ncrna4010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191(3):671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514(7522):389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Galán O, García-Gómez JJ, Kressler D, de la Cruz J. Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae. RNA Biol. 2015;12(8):838–846. doi: 10.1080/15476286.2015.1058477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues CHM, Myung Y, Pires DEV, Ascher DB. mCSM-PPI2: predicting the effects of mutations on protein-protein interactions. Nucleic Acids Res. 2019;47(W1):W338–w344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989. [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16(1):56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. Transcriptome-wide analysis of exosome targets. Mol Cell. 2012;48(3):422–433. doi: 10.1016/j.molcel.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013;38(10):485–493. doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch B, Feigenbutz M, Makino DL, Falk S, Basquin C, Mitchell P, Conti E. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. Embo J. 2014;33(23):2829–2846. doi: 10.15252/embj.201488757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller JM, Falk S, Fromm L, Hurt E, Conti E. Structure of the nuclear exosome captured on a maturing preribosome. Science. 2018;360(6385):219–222. doi: 10.1126/science.aar5428. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater ML. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973;113(1):263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A, Misceo D, Htun S, Mathisen L, Frengen E, Hurtig JE, Enyenihi L, Sterrett MC, Leung SW, Schneidman-Duhovny D, et al. . Biallelic variants in the RNA exosome gene EXOSC5 are associated with developmental delays, short stature, cerebellar hypoplasia and motor weakness. Hum Mol Genet. 2020;29(13):2218–2239. doi: 10.1093/hmg/ddaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somashekar PH, Kaur P, Stephen J, Guleria VS, Kadavigere R, Girisha KM, Bielas S, Upadhyai P, Shukla A. Bi-allelic missense variant, p.Ser35Leu in EXOSC1 is associated with pontocerebellar hypoplasia. Clin Genet. 2021;99(4):594–600. doi: 10.1111/cge.13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJR, Raijmakers R, Pruijn GJM. Dis3-like 1: a novel exoribonuclease associated with the human exosome. Embo j. 2010;29(14):2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterrett MC, Enyenihi L, Leung SW, Hess L, Strassler SE, Farchi D, Lee RS, Withers ES, Kremsky I, Baker RE, et al. . A budding yeast model for human disease mutations in the EXOSC2 cap subunit of the RNA exosome complex. RNA. 2021;27(9):1046–1067. doi: 10.1261/rna.078618.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuparevic I, Mosrin-Huaman C, Hervouet-Coste N, Remenaric M, Rahmouni AR. Cotranscriptional recruitment of RNA exosome cofactors Rrp47p and Mpp6p and two distinct Trf-Air-Mtr4 polyadenylation (TRAMP) complexes assists the exonuclease Rrp6p in the targeting and degradation of an aberrant messenger ribonucleoprotein particle (mRNP) in yeast. J Biol Chem. 2013;288(44):31816–31829. doi: 10.1074/jbc.M113.491290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Nagai K, Akutsu E, Yamaki H, Tanaka N. On the mechanism of action of bleomycin. Strand scission of DNA caused by bleomycin and its binding to DNA in vitro. J Antibiot (Tokyo). 1970;23(10):473–480. doi: 10.7164/antibiotics.23.473. [DOI] [PubMed] [Google Scholar]
- Taylor LL, Jackson RN, Rexhepaj M, King AK, Lott LK, van Hoof A, Johnson SJ. The Mtr4 ratchet helix and arch domain both function to promote RNA unwinding. Nucleic Acids Res. 2014;42(22):13861–13872. doi: 10.1093/nar/gku1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Drazkowska K, Kucinski I, Stodus K, Szczesny RJ, Gruchota J, Owczarek EP, Kalisiak K, Dziembowski A. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 2014;42(2):1270–1290. doi: 10.1093/nar/gkt930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaňáčová Š, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W, et al. . A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3(6):e189. doi:0.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20(2):441–452. doi: 10.1128/MCB.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Yourshaw M, Mamsa H, Rudnik-Schöneborn S, Menezes MP, Hong JE, Leong DW, Senderek J, Salman MS, Chitayat D, et al. . Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat Genet. 2012;44(6):704–708. doi: 10.1038/ng.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511(7510):435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Zinder JC, Zattas D, Das M, Lima CD. Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase. Elife. 2017;6:e29062. doi: 10.7554/eLife.29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick EM, Puno MR, Januszyk K, Zinder JC, DiMattia MA, Lima CD. Helicase-dependent RNA decay illuminated by a Cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell. 2018;173(7):1663–1677 e1621. doi: 10.1016/j.cell.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107(27):12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach S, Langer C, Puppe B, Nedeva T, Bach E, Kull M, Bargou R, Einsele H, Rosenwald A, Knop S, et al. . The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br J Haematol. 2015;169(1):57–70. doi: 10.1111/bjh.13256. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. . Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121(5):725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Yang X, Bayat V, DiDonato N, Zhao Y, Zarnegar B, Siprashvili Z, Lopez-Pajares V, Sun T, Tao S, Li C, et al. . Genetic and genomic studies of pathogenic EXOSC2 mutations in the newly described disease SHRF implicate the autophagy pathway in disease pathogenesis. Hum Mol Genet. 2019;29(4):541–553. doi: 10.1093/hmg/ddz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Lima CD. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31(2):88–100. doi: 10.1101/gad.294769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids summarized in Supplementary Table 1 and Supplementary Table 2 are available upon request. Genomic data from CoMMpass are available at dbGaP with the accession number phs000748.v7.p4. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at G3 online.