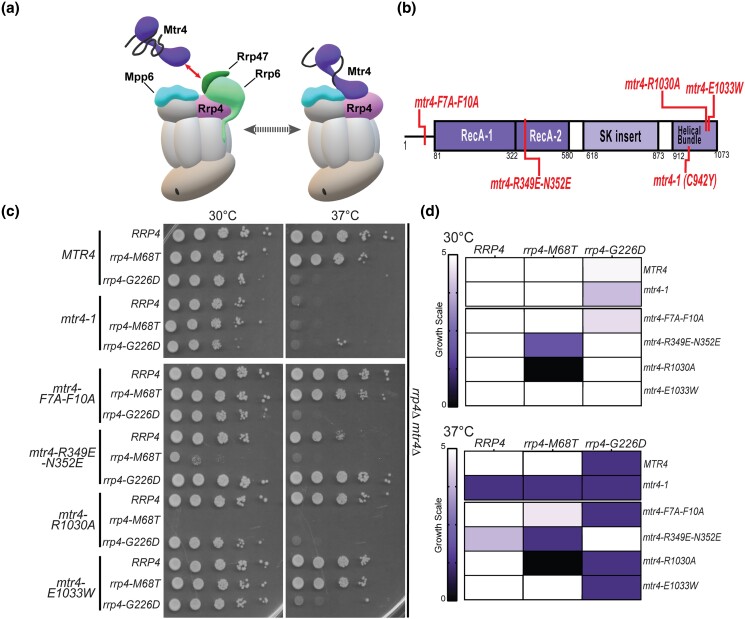
Fig. 6.
The rrp4-M68T mutant cells show specific negative genetic interactions with mtr4 mutants that are predicted to impair the Trf4/5-Air1/2-Mtr4 (TRAMP) complex. a) Cartoon depicting the budding yeast nuclear RNA exosome with interacting nuclear cofactors Mpp6 (turquoise, labeled "Mpp6") and Rrp47 (dark green, labeled "Rrp47"), the exoribonuclease Rrp6 (light green, labeled "Rrp6"), and the essential RNA helicase, Mtr4 (purple) (Schuch et al. 2014; Falk et al. 2017; Schuller et al. 2018). The association of Mtr4 with the RNA exosome is facilitated by interactions between Mtr4 and Rrp6/Rrp47 (denoted by the red arrowed line) and by interactions with Mpp6 which is associated with the Rrp40 RNA exosome subunit and the Rrp4 subunit (Weir et al. 2010; Schuch et al. 2014; Wasmuth et al. 2017). The association of Mtr4 with the RNA exosome can also facilitate interaction with the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex, which triggers degradation of certain RNA targets by adding short oligo(A) tails to the 3′ end of these targets and delivering them to the RNA exosome (Houseley et al. 2006; Anderson and Wang 2009; Belair et al. 2018; Ogami et al. 2018). In addition to Mtr4, the TRAMP complex is composed of a noncanonical poly(A) polymerase, Trf4/5, and a zinc-knuckle RNA binding protein, Air1/2 (Belair et al. 2018). Central to the degradation of TRAMP-targeted RNAs by the RNA exosome is the association of Mtr4 with Trf4/5, Air1/2, and the cap subunits and nuclear cofactors of the RNA exosome complex (Falk et al. 2014; Schuch et al. 2014). b) Domain structure for S. cerevisiae Mtr4. The helicase has a low-complexity N-terminal sequence followed by the conserved helicase region. The helicase region is composed of two RecA domains and a helical domain (labeled helical bundle) that form the globular core typical of DExH family proteins. The helical bundle was originally described as the “ratchet” domain for its role in translocating nucleic acid by a Brownian ratchet (Büttner et al. 2007). In addition, Mtr4 contains an insertion domain and KOW domain that fold into a helical stalk (labeled SK insertion) (Jackson et al. 2010; Weir et al. 2010). The amino acid changes used for this experiment are labeled in red along the domain structure. c) Double mutant cells containing rrp4-M68T and specific mtr4 mutants show lethality at both 30°C and 37°C. The rrp4Δ mtr4Δ double mutant cells were serially diluted, spotted onto solid media, and grown at the indicated temperatures for 3 days. The mtr4 mutant plasmids included in this experiment are as follows; mtr4-1—a temperature-sensitive mutant that contains a missense mutation resulting in the amino acid substitution Cys942Tyr, which causes accumulation of poly(A)+ RNA in the nucleus at 37°C (Kadowaki et al. 1994; Kadowaki et al. 1995; Liang et al. 1996); mtr4-F7A-F10A—an mtr4 allele that impairs the interaction with Rrp6/Rrp47 (Schuch et al. 2014); mtr4-R349E-N352E—a mutation that impairs the association of Mtr4 with the poly(A) RNA polymerase Trf4 with the Mtr4 helicase (Falk et al. 2014); mtr4R-1030A and mtr4-E1033W—two mutations within the helical bundle that differentially impact nucleic acid unwinding by Mtr4 (Taylor et al. 2014). mtr4-1 mutant cells expressing RRP4, rrp4-M68T, or rrp4-G226D show lethality at 37°C presumably due to the known temperature-sensitive nature of the mtr4-1 allele (Liang et al. 1996). Growth of double mutant cells containing rrp4-M68T or rrp4-G226D is shown. d) Summary of rrp4 mtr4 mutant cell growth. Triplicate solid media assays were performed on double mutant cells containing rrp4-M68T or rrp4-G226D and the series of mtr4 variants. Cell growth at both 30°C and 37°C was semiquantified on a scale of zero (no growth; black) to five (comparable to RRP4 wild-type growth; white). Growth scale of the double mutant cells is represented through the color gradient on the two heatmaps. All double mutant cells were generated as described in Materials and Methods. Images shown are from a singular solid media growth assay with all samples plated on the same Leu− media plate. Data are representative of three independent experiments (n = 3).