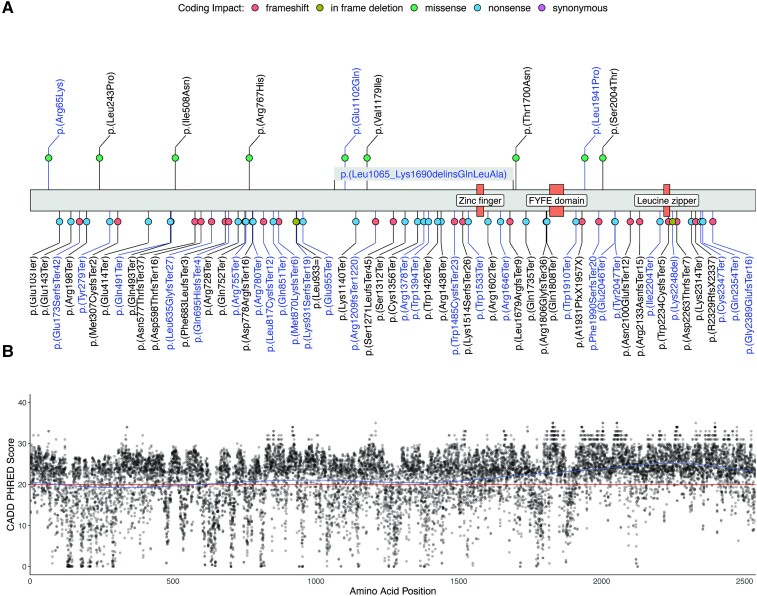
Figure 1.
Molecular spectrum of HSP-ZFYVE26 (SPG15). (A) Schematic of the ZFYVE26 primary protein structure. Novel variants identified in our cohort are labelled in blue, previously reported variants are labelled in black. Coding impacts are colour-coded and missense variants are annotated above the protein structure, while all other variants are depicted below. (B) CADD PHRED scores for all possible missense variants in ZFYVE26 were computed and mapped to the linear protein structure. A generalized additive model was used to predict the tolerance for genetic variation across the protein (blue line). The recommended cut-off value for deleteriousness (CADD PHRED = 20) is marked by a red line.