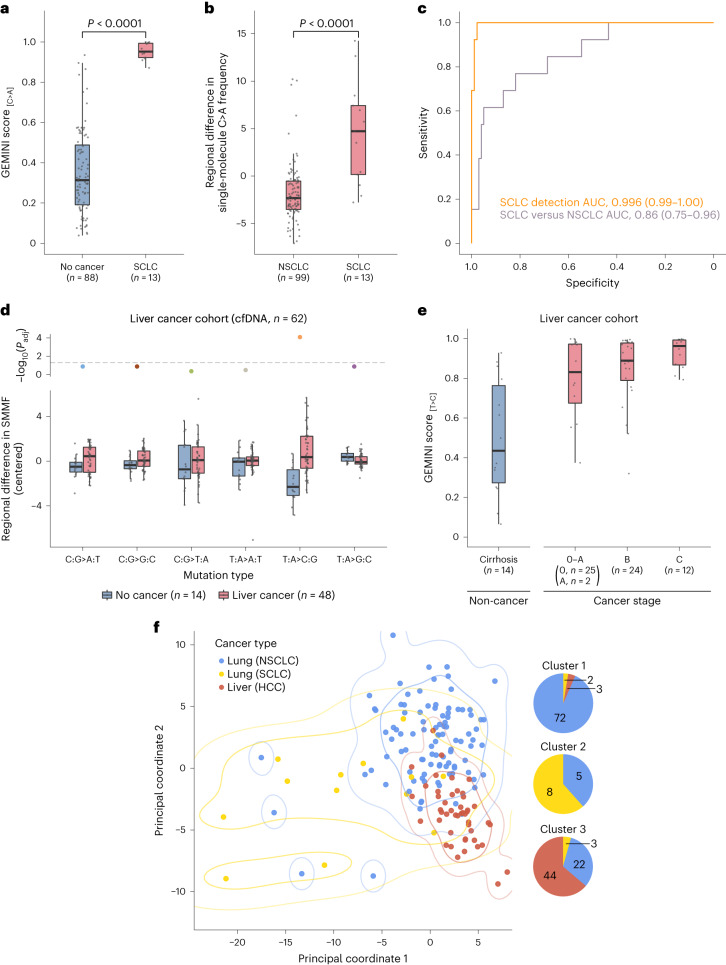
Fig. 5. GEMINI approach for non-invasive detection across multiple cancer types.
a, GEMINI scores in patients with SCLC and high‐risk individuals without cancer in the LUCAS and validation cohorts show high performance for detecting cancer (two-sided Wilcoxon rank sum test, P < 0.0001). b, Regional differences in single-molecule C>A frequency in the LUCAS and validation cohorts demonstrate that GEMINI can be used to identify the bins that are most altered between SCLC and NSCLC (two-sided Wilcoxon rank sum test, P < 0.0001). c, ROC curves for the detection of SCLC (n = 13) compared to non‐cancer controls (n = 88) (orange) as well as for distinguishing SCLC (n = 13) from NSCLC (n = 99) (purple) in the combined LUCAS and validation cohorts. d, Cross-validated regional differences in SMMFs in cfDNA in the liver cancer cohort, median-centered within each mutation type, show a high level of T>C mutations in patients with HCC. Adjusted P values (Padj) were generated using the two-sided Wilcoxon rank sum test and were corrected for multiple comparisons using the Benjamini–Hochberg method. The horizontal dashed line indicates a P value of 0.05. e, GEMINI scores in the liver cancer cohort with the number of individuals indicated at each stage demonstrate high sensitivity for detection of liver cancer across all stages. f, Principal coordinate analysis of the Euclidean distance matrix reflecting cross-validated pairwise differences in regional mutation frequencies between NSCLC, SCLC and HCC. The first two principal coordinates are shown with contours indicating kernel density estimations for 0.7 and 0.95 probability for each cancer type. The composition of cancer types in clusters derived from K-means clustering with k = 3 is indicated to the right. All boxplots represent the interquartile range, with whiskers drawn to the highest value within the upper and lower fences (upper fence, 0.75 quantile + 1.5× interquartile range; lower fence, 0.25 quantile – 1.5× interquartile range). The solid middle line in the boxplot corresponds to the median value.