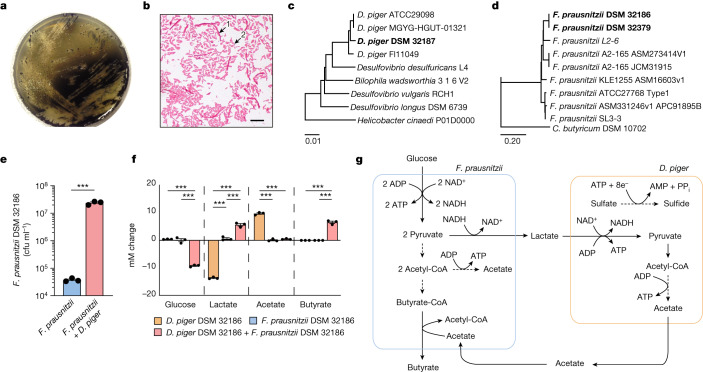
Fig. 1. Co-isolation and cross-feeding of F. prausnitzii and D. piger in vitro.
a, Co-culture of F. prausnitzii DSM 32186 and D. piger DSM 32187 on PGM plates without supplementation of glucose or acetate. b, Gram staining of colonies from isolation of F. prausnitzii DSM 32186 and D. piger DSM 32187. Arrows indicate F. prausnitzii (long fusiform rods) (1) and D. piger (short rods) (2). Scale bar, 10 μm. c, Dendrogram illustrating the relationship between D. piger DSM 32187 and related genomes. d, Dendrogram illustrating the relationship between F. prausnitzii DSM 32186 and related genomes. e, The number of colony-forming units of F. prausnitzii DSM 32186 in monoculture and in co-culture with D. piger DSM 32187 under anaerobic conditions in mPGM (PGM containing 25 mM of glucose) for 24 h. P = 0.0003. f, Metabolite profiles of F. prausnitzii DSM 32186 and D. piger DSM 32187 cultivated as monocultures or co-culture under anaerobic conditions in mPGM medium for 24 h. Glucose: P = 0.0000031 (F. prausnitzii + D. piger versus D. piger), P = 0.0000038 (F. prausnitzii + D. piger versus F. prausnitzii); lactate: P = 0.0000001 (F. prausnitzii + D. piger versus D. piger), P = 0.0000005 (F. prausnitzii versus D. piger), P = 0.00014 (F. prausnitzii + D. piger versus F. prausnitzii); acetate: P = 0.0000004 (F. prausnitzii + D. piger versus D. piger), P = 0.0000003 (F. prausnitzii versus D. piger); butyrate: P = 0.000001(F. prausnitzii + D. piger versus D. piger), P = 0.0000031 (F. prausnitzii + D. piger versus F. prausnitzii). ‘mM change’ on the y axis indicates the difference in concentration from the inoculated medium at baseline. g, Schematic of the suggested cross-feeding between F. prausnitzii and D. piger as co-culture in mPGM. n = 3 independent experiments, ***P < 0.001 determined by two-tailed t-test (e) or one-way ANOVA (f). Data are mean ± s.e.m.