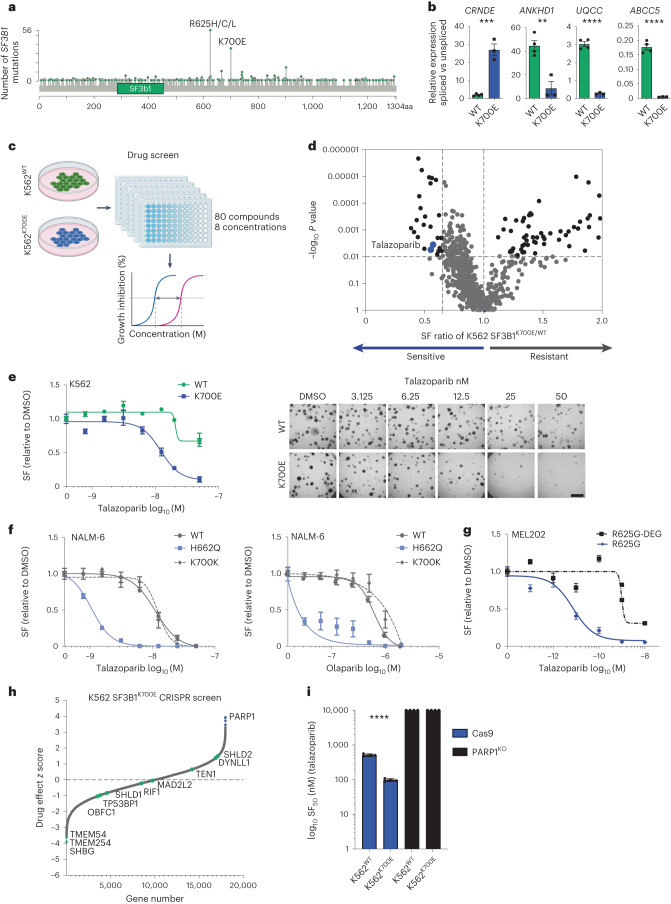
Fig. 1. SF3B1 hotspot mutations lead to PARPi sensitivity in isogenic models.
a, Lollipop plot of the number of SF3B1 mutations in TCGA (pan-cancer cohort and MSK IMPACT clinical sequencing study (n = 21,912). Data from cBioportal. b, qRT–PCR of differentially spliced exons of selected indicator genes in the myeloid leukemia isogenic cell lines (K562) that express wild-type (WT) or mutant (K700E) SF3B1 (n = 3 independent biological replicates). Data are mean ± s.e.m., unpaired two-tailed t-test; CRNDE, P = 0.0003; ANKHD1, P = 0.0036; UQCC, P < 0.0001 and ABCC5, P < 0.0001. c, Schematic of small-molecule inhibitor screening pipeline. d, Volcano plot of compound selectivity from the small-molecule inhibitor library screen in K562 cell lines (−log10 P < 0.01 unpaired two-tailed t-test and surviving fraction (SF) ratio K562 SF3B1K700E/SF3B1WT < 0.6). Blue dots indicate two independent concentrations of the PARPi talazoparib. e, Fourteen-day clonogenic dose–response curves and representative images of K562 isogenic cells harboring the K700E SF3B1 hotspot variant and wild-type cells following exposure with the PARPi talazoparib (scale bar = 4 mm). f, Fourteen-day clonogenic dose–response curves of NALM-6 isogenic cells with the H662Q SF3B1 hotspot variant, K700K silent variant and wild-type cells following exposure with talazoparib and olaparib (n = 3 independent biological replicates, error bars show ± s.e.m.) g, Fourteen-day clonogenic dose–response curves of uveal melanoma MEL202R625G cells with the endogenous R625G SF3B1 hotspot variant, and revertant MEL202R625G-DEG cells following exposure with talazoparib. Data are mean normalized to DMSO control from n = 3 independent biological experiments, error bars show ± s.e.m (e–g). h, Waterfall plot of whole-genome CRISPR screen in K562 SF3B1K700E cells, depicting hits (blue) from n = 3 independent biological replicate experiments. Genes known to cause resistance to PARPi in homologous recombination-deficient cells are highlighted. i, Bar plot depicting the SF50 (concentration of drug that allows 50% cell survival) values of K562 SF3B1 wild-type and K700E cells with Cas (control) or CRISPR PARP1KO under talazoparib exposure (n = 3 independent biological repeats). Error bars show mean ± s.e.m. Unpaired two-tailed t-test, Cas9 wild-type versus K700E. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (b,i). SF, surviving fraction.