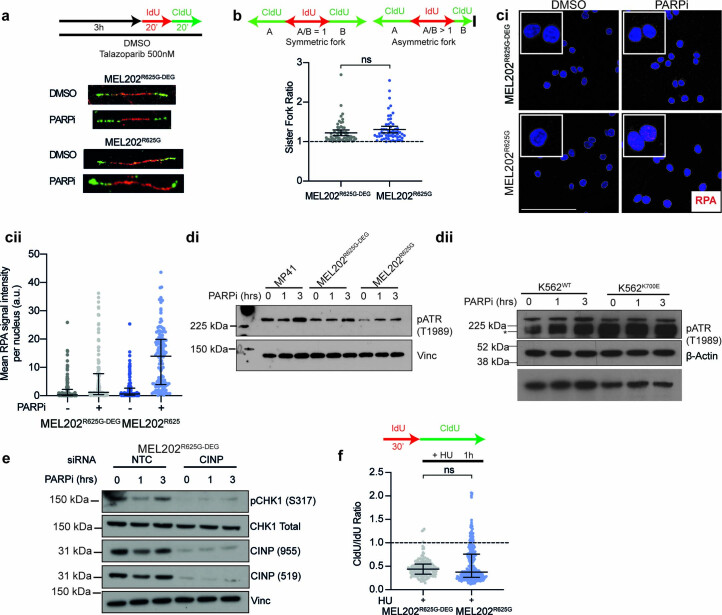
Extended Data Fig. 5. SF3B1 mutant cells elicit a defective RS response under PARPi.
a, Experimental set up of fiber assay and representative immunofluorescence images of IdU and CldU labeled DNA fibers after 3 hours 500 nM talazoparib or DMSO exposure. b, Schematic of analysis and scatterplot of quantification of sister fork ratio taken from DNA fiber analysis of MEL202 isogenic cells exposed to DMSO. Data are mean of n = 3 biological replicates, error bars show ± s.e.m. (unpaired two-tailed t-test (NS P = 0.1337)). c, Representative immunofluorescence images (ci) and scatterplot (cii) of RPA foci in MEL202 isogenic cells following 3 hours of 500 nM talazoparib or DMSO exposure. Data are from n = 2 biological replicates, error bars show ± s.d. of foci in individual nuclei. Scale bar = 100 μm. d, Western blot of pATR (T1989) in MP41WT and MEL202 isogenic cells (di) and K562 isogenic cells (dii) at 0, 1, or 3 hours of 500 nM talazoparib exposure. e, Western blot of pCHK1 (S317), total CHK1, and CINP expression using two different CINP antibodies in MEL202R625G-DEG cells after non-targeting control (NTC) or CINP siRNA gene mediated silencing, at 0, 1, or 3 hours of 500 nM talazoparib exposure (n = 1 biological replicate). f, Scatterplot of CldU/IdU ratio taken from DNA fiber analysis of MEL202 isogenic cells exposed to 100 μM hydroxyurea (HU). Data are mean of n = 3 biological replicates, error bars show ± s.e.m. (unpaired two-tailed t-test (NS P = 0.458).