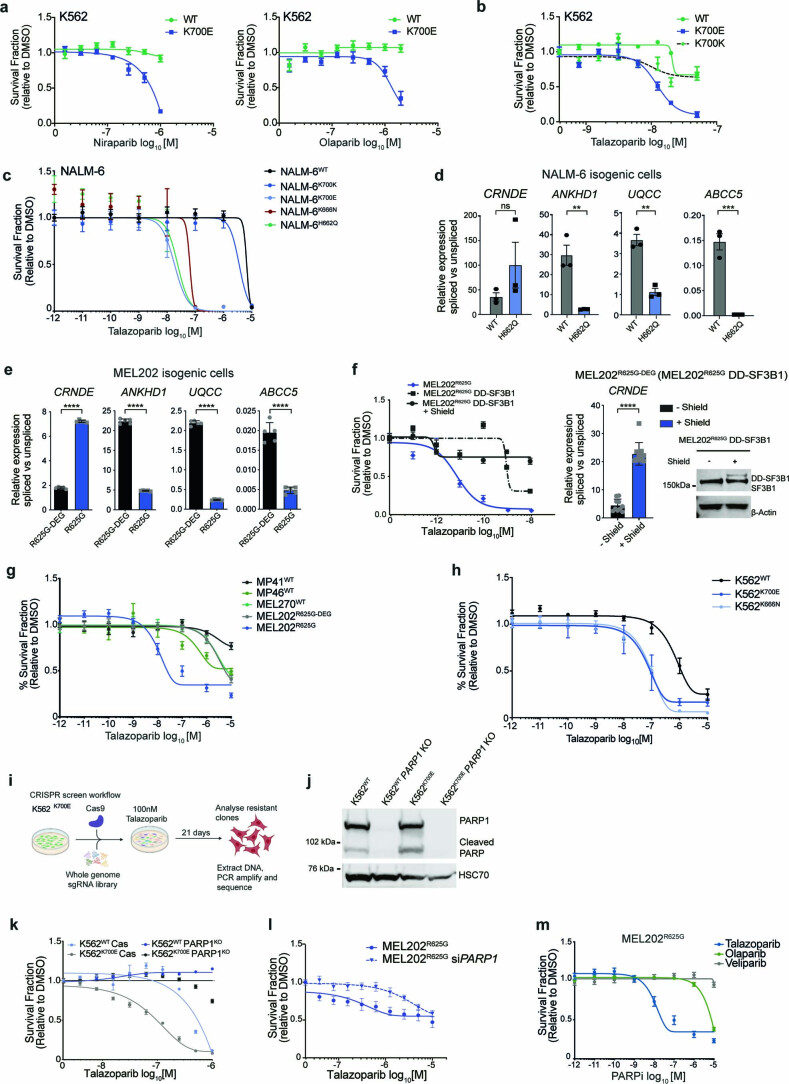
Extended Data Fig. 1. SF3B1 hotspot mutations induce mis-splicing and PARPi sensitivity.
a, b, 14 day clonogenic dose–response curves of K562 SF3B1WT, SF3B1K700K (silent mutation) and SF3B1K700E isogenic cells following exposure with distinct PARP inhibitors. Data are mean ± s.e.m, (n = 3 independent biological replicates). c, 14 day (3D viability) talazoparib dose–response curves of NALM6WT, NALM6K700K and SF3B1MUT NALM6K700E, NALM6K666N and NALM6H662Q cell lines grown as spheroids. Data are mean ± s.e.m, (n = 3 independent biological replicates). d, Representative qRT-PCR of differentially spliced exons of indicator genes in the NALM-6WT and NALM-6H662Q isogenic lines. Data are mean of n = 3 biological replicates, ± s.d. (unpaired two-tailed t-test (NS P = 0.2426, **P = 0.0058, **P = 0.0015, ***P = 0.0009)). e, Representative qRT-PCR of differentially spliced exons of indicator genes in the MEL202R625G-DEG and MEL202R625G cells. Data are mean of n = 5 biological replicates, ± s.d. (unpaired two-tailed t-test (****P < 0.0001)). f, 14 day clonogenic dose–response for the isogenic MEL202R625G-DEG and MEL202R625G cells exposed to talazoparib and revertant MEL202R625G-DEG cells labeled with a degron tag (MEL202R625G DD-SF3B1) +/- Shield-1 compound to stabilize expression of the mutant allele. Data are normalized to DMSO control and presented as mean ± s.e.m. (n = 3 biological replicates). qRT-PCR of differentially spliced exon of CRNDE in the MEL202R625G-DEG +/- shield compound (n = 3 biological replicates, ****P < 0.0001, unpaired two-tail t-test). Western blot analysis of MEL202R625G-DEG (MEL202R625G DD-SF3B1) showing protectable mutant allele upon shield compound treatment. g, 5-day viability dose–response curves of wild-type uveal melanoma cell lines MP41, MP46, MEL270 and MEL202. Data are mean ± s.e.m. (n = 3 biological replicates). h, 14 day (3D viability) dose–response curves of K562WT, K562K700E and K562K666N spheroids exposed to talazoparib. Data are mean ± s.d. (n = 3 independent biological replicates). i, Schematic of CRISPR screen workflow. j, Western blot of PARP1, cleaved PARP1 and HSC70 in K562WT and K562K700E cells with Cas or PARP1 KO. k-l, Talazoparib dose-response curves showing the survival fraction of K562 isogenic cells +/- PARP1 CRISPR knockout (KO) (k), MEL202R625G cells +/- PARPi siRNA (l). Data are mean of n = 3 biological replicates, ± s.e.m. m, 5 day dose–response curve of MEL202R625G cells exposed to talazoparib, olaparib and veliparib. Data are mean ± s.e.m. (n = 3 biological replicates).