Fig. 3.
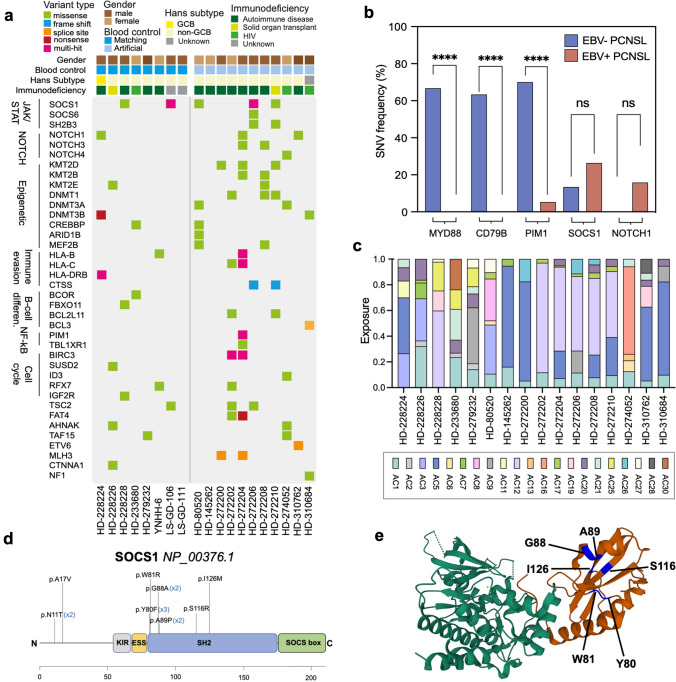
Single nucleotide variants in EBV+ PCNSL. a Filtered genes with functional exonic SNVs and INDELs are shown. Colors indicate the type of alteration. SOCS1 was the most frequently (26%, n = 5) altered gene in EBV+ PCNSL. Mutually exclusive NOTCH variants were detected in 26% (n = 5) of tumors. b Frequencies of selected alterations are contrasted to EBV− PCNSL (blue) from the ICGC-MMML-Seq dataset. EBV+ cases (red) mostly lacked MYD88, CD79B, and PIM1 variants characteristic of virus-negative disease (blue). Frequencies were compared with Fisher Exact tests and significance levels are displayed (****p < 0.0001; ns = not significant). c Normalized single base substitution Alexandrov-COSMIC (AC) mutational signatures in EBV+ PCNSL are shown. Colors indicate the signature type and proportions reflect related SNVs counts/sample. Signatures associated with spontaneous deamination of cytosines (AC1), and defective DNA mismatch repair (AC20, AC21) were frequently found. d The SOCS1 gene and its domains are displayed (KIR, kinase inhibitory region; ESS, extended SH2 subdomain; SH2, Src2 homology domain; SOCS box) and detected alterations are indicated. Most (n = 10/13) variants clustered to the SH2 domain, which mediates binding and subsequent inhibition of JAK1. e Crystal structure of the SOCS1 SH2 domain (red) in complex with the JAK1 kinase domain (green) obtained from the RCSB protein data bank (accession number 6C7Y). Positions of altered amino acids (blue) in EBV+ PCNSL are indicated within the complex