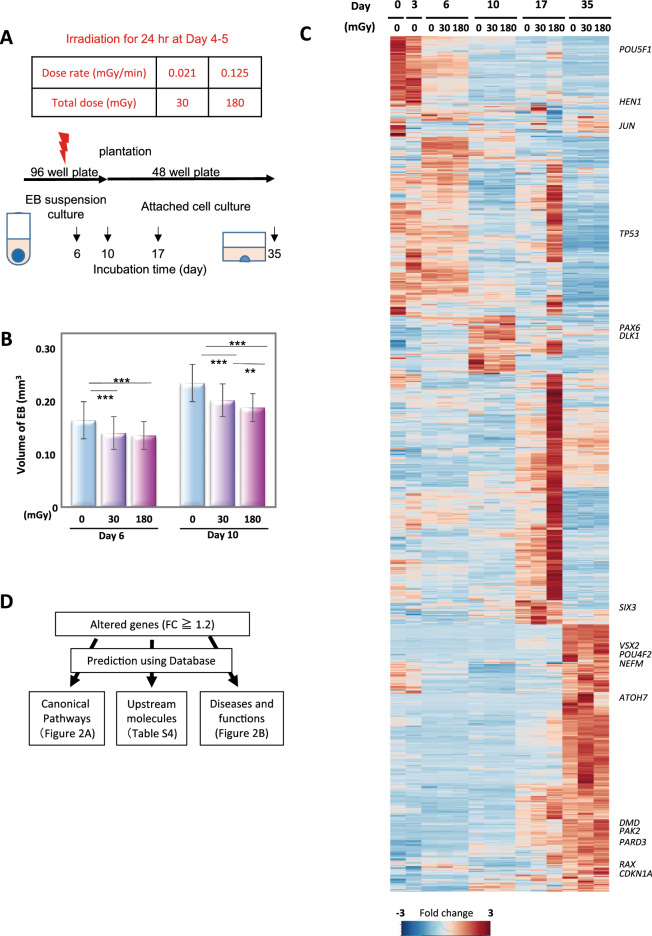
Figure 1.
Growth suppression of EB and altered gene expression by low-dose radiation. (A) Doses and time course of the exposure experiment. On day 0, iPSCs were placed into 96-well culture plates. After EB formation, the cells were irradiated from days 4 to 5. The doses (dose rates) were 30 mGy (0.021 mGy/min) and 180 mGy (0.125 mGy/min). On day 10, the cells were transferred to 48-well flat bottom culture plates coated with iMatrix-511. On days 0, 3, 6, 10, 17, and 35, RNAs were analysed. (B) Growth suppression by low-dose radiation. EB sizes are shown. Data are presented as the mean and standard deviation of 10 EBs. Experiments were repeated three times on different days. ∗∗∗p < 0.001, ∗∗p < 0.01 (Student’s t test). Error bar: average deviation of three experiments. (C) Clustering and heatmap of RNA-seq data. RNA including six to twelve EBs at each point was isolated. Of the 26,260 genes, 5,246 genes had > 25 FPKM that met one or more conditions (Supplemental Table S1). Among them, 1,007 genes with changes in expression ≥ 1.4-fold compared with the FPKM for nonirradiated cells on the same day at least once were selected (Supplemental Table S2). After selection, the FPKM of each gene was normalized and clustered. RNA-seq was performed once for days 1, 3, 10, and 17 and three times for days 6 and 35. Each RNA-seq sample included six to twelve EBs. (D) IPA schematic. Radiation-induced changes in gene expression were examined by IPA in three independent ways. First, relevant “upstream molecules” were predicted by summarizing the altered genes matched with downstream genes. Second, altered genes were matched to registered genes in each “canonical pathway.” Third, downstream “Diseases and functions” were predicted by matching the altered genes with genes in the database. For all “canonical pathways,” “upstream molecules,” and “diseases and functions,” P values and Z scores were calculated. The P value is the calculated significance using Fisher’s exact test (calculated as − log p). The Z score is the predicted degree of activation (positive) or inhibition (negative).