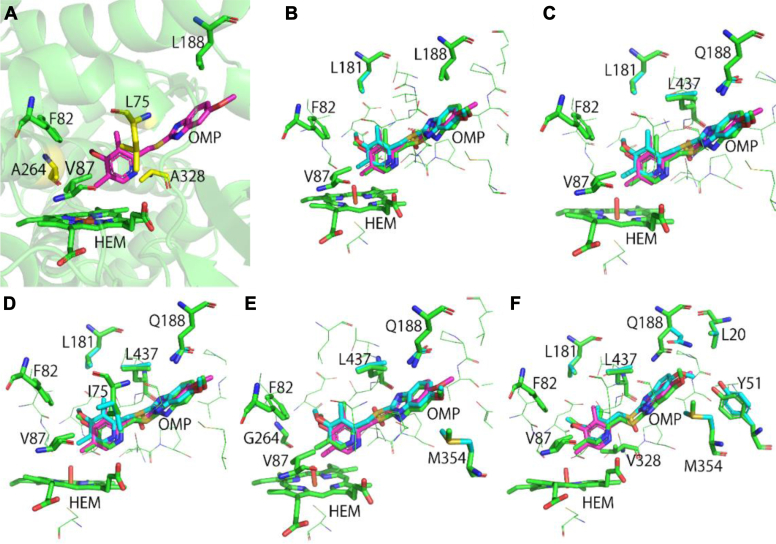
Figure 2.
Computational design and modeling with UniDesign.A, locations of main design sites. Mutable sites L75, A264, and A328 are colored yellow, whereas F82, V87, L188, and HEM are in green. Ligand OMP is colored in magenta. Their coordinates were obtained from the Protein Data Bank (PDB ID: 4KEY). It is of note that the sulfone oxygen is missing from the crystal structure because of weak electron density. B, UniDesign model of the A82F/F87V double mutant (DM). The UniDesign-generated (R)- and (S)-OMP poses are colored green and cyan, respectively. The crystal OMP (in magenta) is depicted for comparison. The main design sites and other sites that adopt different conformations for binding (R)- and (S)-OMP are shown in green and cyan sticks, respectively. The color scheme is the same in B–F. C, UniDesign model of the A82F/F87V/L188Q triple mutant (TM). D, UniDesign model of the UD1 mutant (TM/L75I). E, UniDesign model of the UD2 mutant (TM/A264G). F, UniDesign model of the UD3 mutant (TM/A328V). OMP, omeprazole.