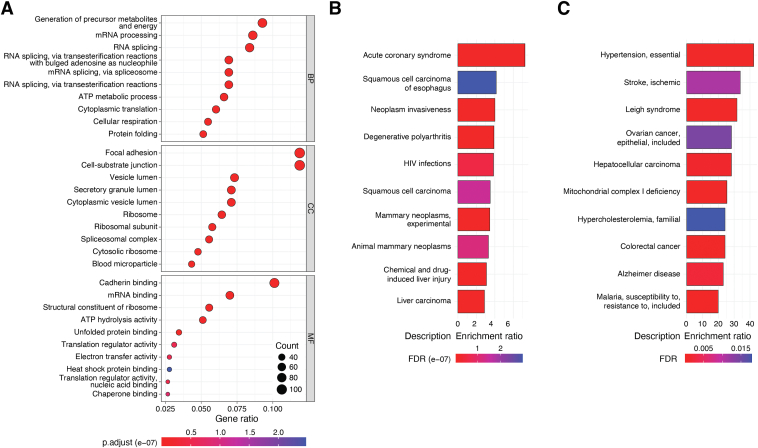
Figure 3.
Analysis of tyrosine-nitrated proteins.A, functional analysis of the 908 tyrosine-nitrated proteins using the gene ontology data base. The data depicts the top ten annotations for biological processes (BP), cellular component (CC), and molecular function (MF) considering the values for false discovery rate (values are indicated by the heat map) and fold enrichment. The size of the circle indicates the number of genes annotated in the corresponding BP, CC, and MF. B, bioinformatic analysis of the 908 tyrosine-nitrated proteins for association (over-representation) with human disorders, using DisGeNET, a collection of genes and gene variants associated with human disease or (C), the online catalog of Human Genes and Genetic Disorders (https://www.omim.org). The top ten human associated disorders based on the false discovery rate (FDR) values are shown.