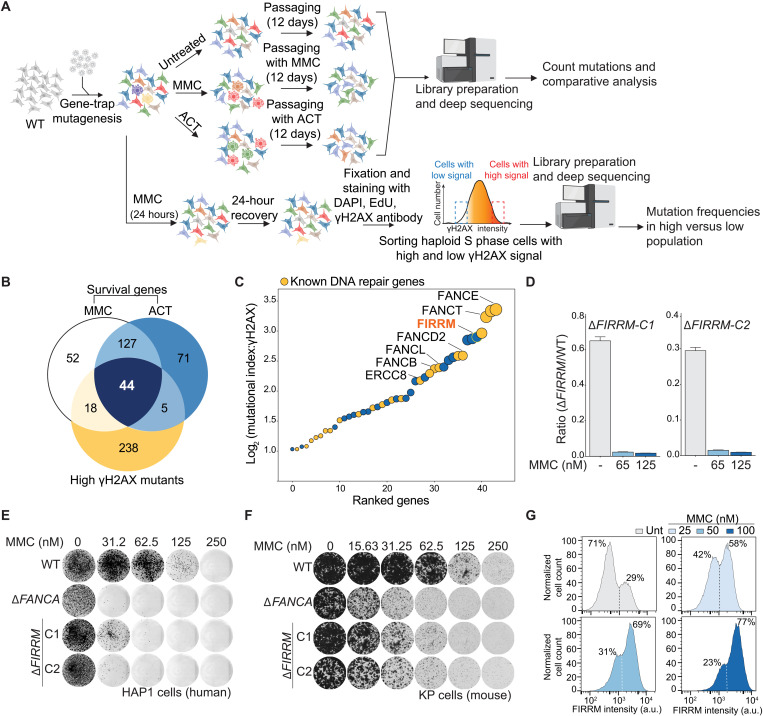
Fig. 1. Haploid genetics identify FIRRM as an important factor for genome stability in response to ICLs.
(A) Schematic overview of the haploid genetic screens performed using survival or γH2AX staining as readouts. (B) Venn diagram of the genes identified by the three genetic screens. (C) Ranking plot depicting the 44 genes commonly identified between the performed genetic screens. Genes are ranked on the basis of the mutational index of the γH2AX screen. Orange dots indicate known DNA repair genes, and blue dots indicate genes with an unknown function in ICL repair. (D) Competition growth assay with wild-type (GFP) and ΔFIRRM (mCherry) HAP1 cells treated with MMC (65 or 125 nM) for 8 days. Data are represented as mean ratio (mCherry/GFP) ± SEM. (E and F) Clonogenic survival in response to different concentrations of MMC in HAP1 cells (E) and mouse mammary tumor cells (K14Cre;Trp53−/−; KP) (F). WT, wild type. (G) HAP1 cells carrying an endogenous V5-tag at the C terminus of FIRRM were transduced with a 1:1 mix of nontargeting sgRNA and a sgRNA targeting FIRRM. After puromycin selection, the pool of cells was treated with different concentrations of MMC for 7 days. Samples were then fixed, stained with a V5 antibody, and analyzed by flow cytometry. All experiments were performed in triplicates except the genetic screens. For the survival-based screens, two biological replicates were performed, and for the γH2AX screen, one replicate was done. a.u., arbitrary unit.