FIGURE 1.
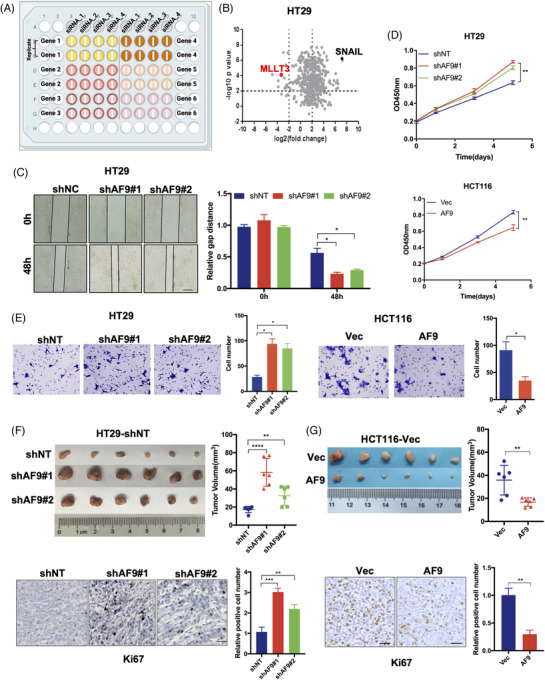
siRNA screening of epigenetic regulator factors (ERFs) discovers AF9 plays a suppressive role of colorectal cancer (CRC). (A and B) HT29 cells were transfected with individual siRNA targeting 591 ERFs (four siRNA for each gene) or siRNA targeting SNAI1 as the positive control or non‐targeting control siRNA (siNC) as the negative control, respectively. Scratch wound healing assays were performed. Real‐time gap distances were measured using IncuCyte high‐throughput screening system for 48 h. Schematic diagram of screening strategy was presented (A). Data presented as volcano plot of two independent experiments (B, top panel). The small interfering RNAs (siRNAs) with p value < .01 and migration distance (normalised to siNC) < .25 or > 4 was considered to be effective siRNAs that significantly influence tumour cell activities. ERFs targeted by more than two effective siRNAs were selected as candidate ERFs required for tumour cell activities (B). (C) Representative images of wound healing assays performed in HT29 cells with or without shAF9 were shown at indicated time point (scale bar represents 400 μm). (D) Cell proliferation of HT29 cells with or without shAF9, or HCT116 cells with or without AF9 overexpression was measured by CCK‐8. (E) Representative images of transwell assays performed in HT29 cells with or without shAF9 or in HCT116 cells with or without AF9 overexpression were shown (scale bar represents 60 μm). (F and G) Xenograft formation. HT29 cells with or without AF9 or HCT116 cells with or without AF9 overexpression were implanted into left groin of nude mice (n = 6 for each group). The tumour volume was measured at the end of the experiment. Ki67 staining was performed to test the in vivo proliferation (*p < .05, **p < .01, ***p < .001, ****p < .0001).
