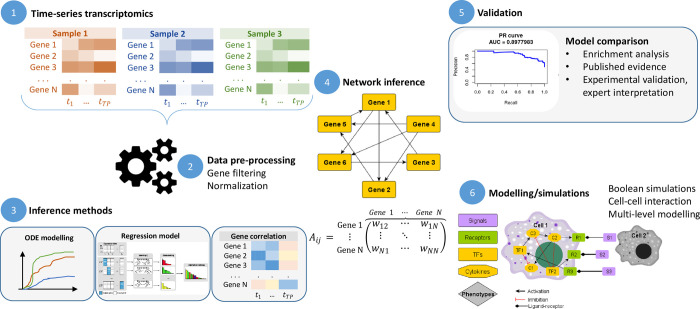
Fig 2. From time-series transcriptomics to GRN inference and model validation.
The process usually starts with processing the time-series datasets and identifying the time-points that are relevant to describe the biological process (1). As a second step, data formatting, normalization and gene filtering and/or binarization might be necessary (2), depending on the inference method to be used (3). The inferred network consists of a weighted directed GRN (4). Model validation (5) is then necessary to quantify the method performance in inferring the GRN when compared to gold-standard datasets, evidence from literature or prior biological knowledge. The inferred network can further be used for the application of dynamical models in cellular or multilevel modeling (6).