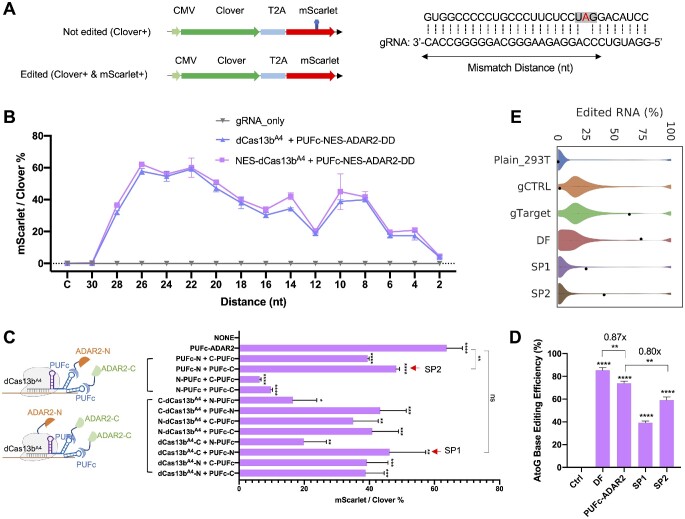
Figure 2.
CREST-mediated A-to-G base editing. (A) Left: schematic of the reporter system for A-to-G editing. CMV-driven Clover and mutant mScarlet ORFs were separated by a T2A self-cleaving peptide. Before editing, the premature stop codon (UAG) within the downstream mScarlet ORF disrupts its complete translation, marking transfected cells Clover-positive only (top). A-to-G editing converts UAG to UGG and therefore rescues the translation of the complete mScarlet protein, turning cells double positive for Clover and mScarlet (bottom). Right: target sequence within the mutant mScarlet (top sequence) and spacer sequence in gRNA (bottom sequence). Mismatch distance is defined as the distance between first matched nucleotide to the target adenine as indicated by the double arrow. (B) Tilling of mismatch distances of A-to-G editing gRNAs from 2nt to 30nt. HEK293T cells were co-transfected with reporter, gRNA with or without dCas13bA4, PUFc-ADAR2-DD, as indicated. Flow cytometry was done 48 hours after transfection and the editing efficiency was calculated by the ratio of the percentages of mScarlet- and Clover-positive cells (Y axis). X-axis: the mismatch distance of spacer sequence in gRNAs. C stands for non-targeting control gRNA. Different transfection groups are shown as indicated at the right. All gRNAs were tagged by 10xPBSc-Loop. (C) Reconstitution of split ADAR2-DD by CREST system. ADAR2-DD was split into two fragments (N terminal and C terminal) and fused with PUFc (Top left) or dCas13A4 and PUFc separately (Bottom left). X-axis: Editing efficiency measured by flow cytometry. Y-axis: different orientations and combinations of fusion proteins are indicated. NONE: cells transfected with gRNA only were used as the negative control. The intact ADAR2-DD fused with PUFc (PUFc-ADAR2) was used as a positive control. N stands for the N-terminal fragment of ADAR2-DD and C stands for the C-terminal fragment of ADAR2-DD. All gRNAs were tagged by 3xPBSc-Loop with 22nt mismatch distance. (D) A-to-G editing efficiency quantified by Sanger sequencing. Ctrl: HEK293T cell transfected with reporter, dCas13A4, PUF-ADAR2-DD and the non-targeting control gRNA. DF: direct fusion of ADAR2-DD with original dCas13b without AAAA mutation and the on-target gRNA without RNA scaffolds. PUFc-ADAR2: HEK293T cells transfected with reporter, dCas13A4, PUF-ADAR2-DD and the on-target gRNA tagged by 3xPBSc-Loop. SP1 and SP2 are indicated in (C). (E) Violin plots representing distribution of A-to-G editing yields observed at reference sites. Plain HEK293T cells without transfection was used as control and the editing yield at the target site was manually set to zero. Effectors transfected into HEK293T cells were as indicated and black dots indicate editing yields at the target site within the mScarlet transcript. Three replicates were done for each group. Data were displayed as mean ± SD, n = 3. *P< 0.05, ** P < 0.01,***P< 0.001, ****P< 0.0001, ns, not significant, by two-sided t-test.