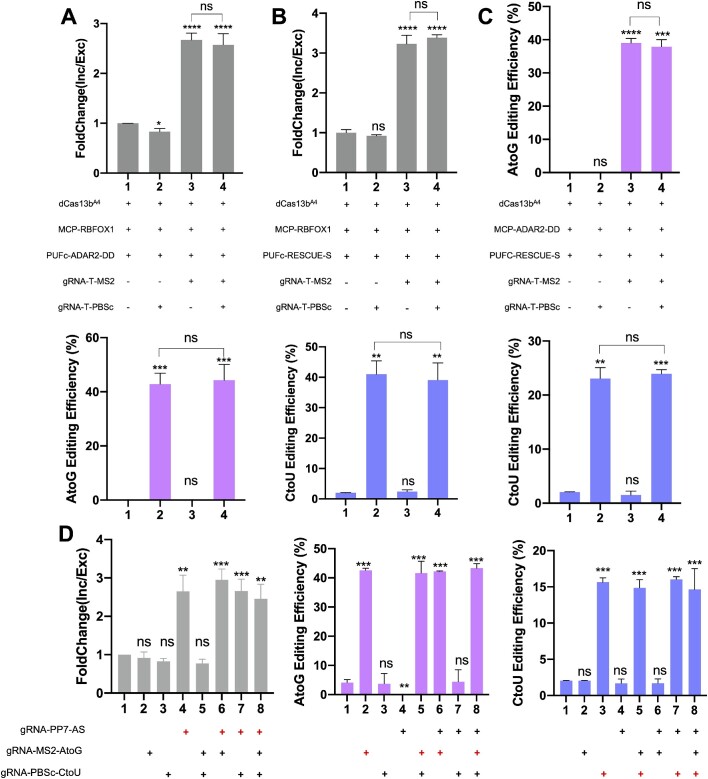
Figure 5.
CREST-mediated multifunctional RNA modulation. (A-–C) HEK293T cells were co-transfected with dCas13bA4 and MCP/PUFc-fused effectors as indicated in the middle. Desired modulations were achieved by addition of gRNAs with effector-matched MS2, PBSc, or PP7 scaffolds. (A) Simultaneous induction of alternative splicing by gRNA-1xMS2 and A-to-G base editing by gRNA-3xPBSc-Loop. Top: Inclusion of exon 7 in SMN2 transcript measured by RT-qPCR. Middle: The design of transfection groups. Bottom: A-to-G editing efficiency quantified by Sanger sequencing (n = 3). (B) Simultaneous induction of alternative splicing by gRNA-1xMS2 and C-to-U base editing by gRNA-10xPBSc-Loop. Top: Inclusion of exon 7 in SMN2 transcript measured by RT-qPCR. Middle: The design of transfection groups. Bottom: C-to-U editing efficiency quantified by sanger sequencing (n = 3). (C) Simultaneous A-to-G base editing by gRNA-1xMS2 and C-to-U based editing by gRNA-10xPBSc-Loop. A-to-G (top) and C-to-U (bottom) editing efficiency quantified by Sanger sequencing (n = 2). (D) HEK293T cells were co-transfected with dCas13bA4, PCP-RBFOX1, MCP-ADAR2-DD, PUFc-RESCUE-S for splicing, A-to-G, C-to-U editing, respectively. Desired modulations were achieved by addition of gRNAs with effector-matched MS2, PBSc or PP7 scaffolds as indicated at the bottom. Left: Fold change of inclusion of exon 7 in SMN2 minigene quantified by RT-qPCR. Middle and right: A-to-G and C-to-U editing efficiency on reporters measured by Sanger sequencing. Data were displayed as mean ± SD. The first column in each graph was used as the control to which all other columns were normalized. *P< 0.05, ** P < 0.01,***P< 0.001, ****P< 0.0001, ns, not significant, by two-sided t-test.