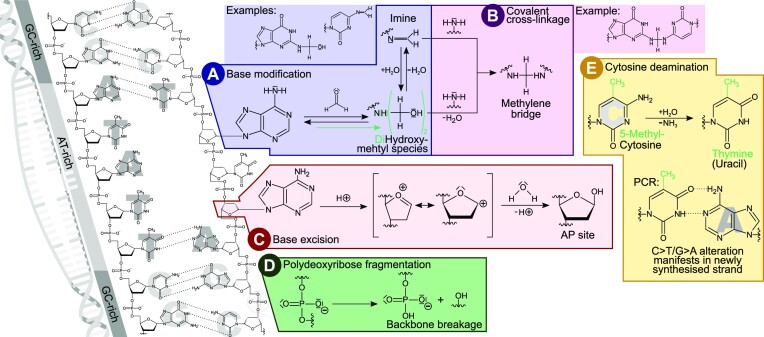
Figure 1.
Summary of DNA modifications typically observed in FFPE samples. DNA instability is initiated by double strand denaturation and base unstacking, especially in AT-rich regions (far left). Modifications influencing base pairing then induce further local double strand denaturation and accelerate base modifications, leading to local hot spots of alterations. (A) Base modification caused by the nucleophilic attack of a base's amino group towards the electrophilic carbon of formaldehyde. The resulting hydroxymethyl can condensate to form an imine (altering base pairing) or further react to a dihydroxymethyl species. (B) Methylene bridges can form a covalent crosslink with another nucleophilic group of, e.g. a base or a protein, both leading to DNA polymerase blockage. (C) Base excision by hydrolysis of the N-glycosylic bond leaves a 2-deoxy-d-ribose AP site in the phosphate backbone. A transition state can form as an intermediate containing a highly reactive cyclic oxocarbenium ion that reacts with water. (D) Formaldehyde conservation also promotes the slow hydrolysis of phosphodiester bonds that breaks the phosphate backbone and fractures the DNA. (E) As glycosylase repair enzymes are inactivated by the fixation, spontaneous cytosine deamination converting cytosine to uracil is no longer corrected. In case of 5-methylcytosine this conversion results in thymine. Either way, the base will now pair with adenine instead of the original C/G base pair at that location.