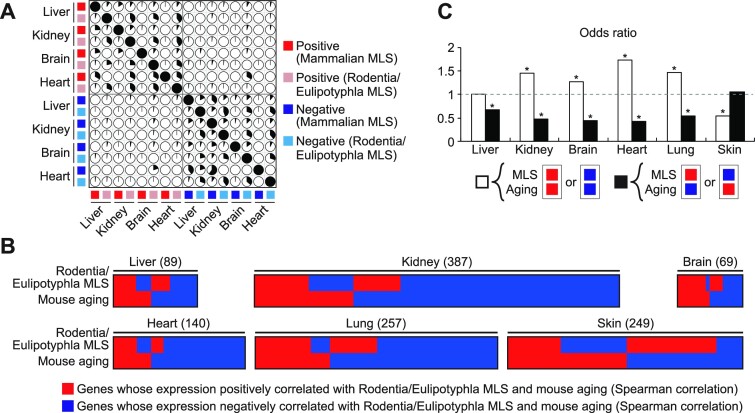
Figure 3.
A significant similarity between gene expression patterns associated with Rodentia/Eulipotyphla MLS and mouse aging. (A) The correlation plot represents the overlap among mammalian and Rodentia/Eulipotyphla MLS-associated genes identified in liver, kidney, brain, and heart. Each pie represents the percentage of overlapping genes in the gene set shown on the left side. (B) The heatmaps show the overlap between genes associated with Rodentia/Eulipotyphla MLS and mouse aging. Each column represents a single gene and the total number of genes shown in the heatmap is indicated in the parenthesis. The top rows show whether the gene was positively (red) or negatively (blue) associated with Rodentia/Eulipotyphla MLS in the tissue shown on the top of the heatmap. The bottom rows indicate whether the gene was upregulated (red) or downregulated (blue) during aging in mouse in the tissue shown on the top of the heatmap. Only MLS-associated genes whose expression was also affected by aging are shown in the heatmaps. (C) Bar graphs represent the odds ratios of concordant (white bars) and discordant genes (black bars). * FDR < 0.05 (binominal test).