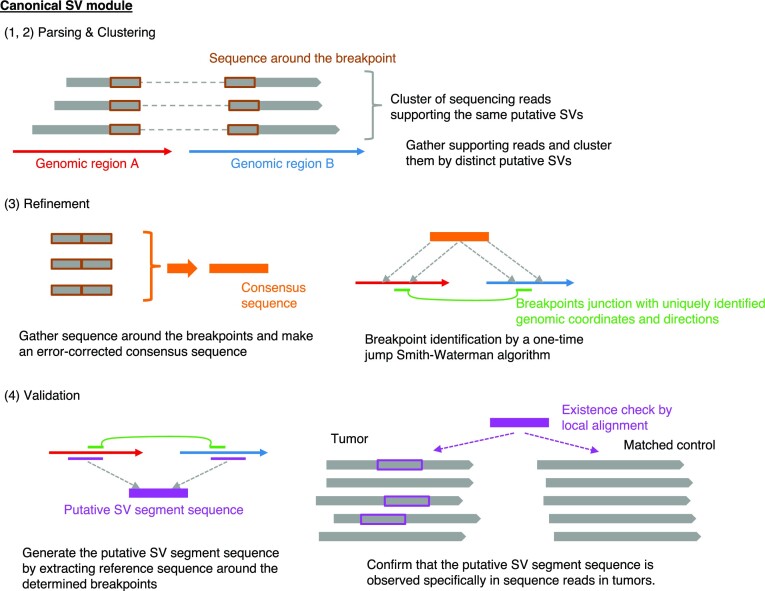
Figure 1.
Workflow of somatic SV detection in nanomonsv Canonical SV module. Canonical SV module for nanomonsv consists of the following four steps. Parsing: the reads likely supporting SVs are extracted from both tumor and matched control BAM files using CIGAR string and supplementary alignment information. Clustering: the reads from the tumor sample that presumably span the same SVs are clustered, and the possible ranges of breakpoints are inferred for each possible SV. If there exist apparent supporting reads in the matched control sample (or non-matched control panel samples when they are available), these are also removed. Refinement: Extract the portions of the supporting reads around the breakpoints, and perform error-correction using racon (78) to generate a consensus sequence for each candidate SV. Then, aligning the consensus sequence to those around the possible breakpoint regions in the reference genome using a modified Smith-Waterman algorithm (which allows a one-time jump from one genomic region to the other, see Supplementary Figure S1), we identify the exact breakpoint positions and the inserted sequence inside them. Validation: From the breakpoint determined in the previous step, we generate the ‘putative SV segment sequence.’ Then we collect the reads around the breakpoint of putative SVs and check whether the putative SV segment sequence exists (then the read is set as a ‘variant supporting read’) or not (then the read is classified to a ‘reference read’) in each read of the tumor and matched control. Finally, candidate SVs with ≥3 variants supporting reads in the tumor and no variant supporting reads in the matched control sample are kept as the final SVs. See Supplementary Text for detail.