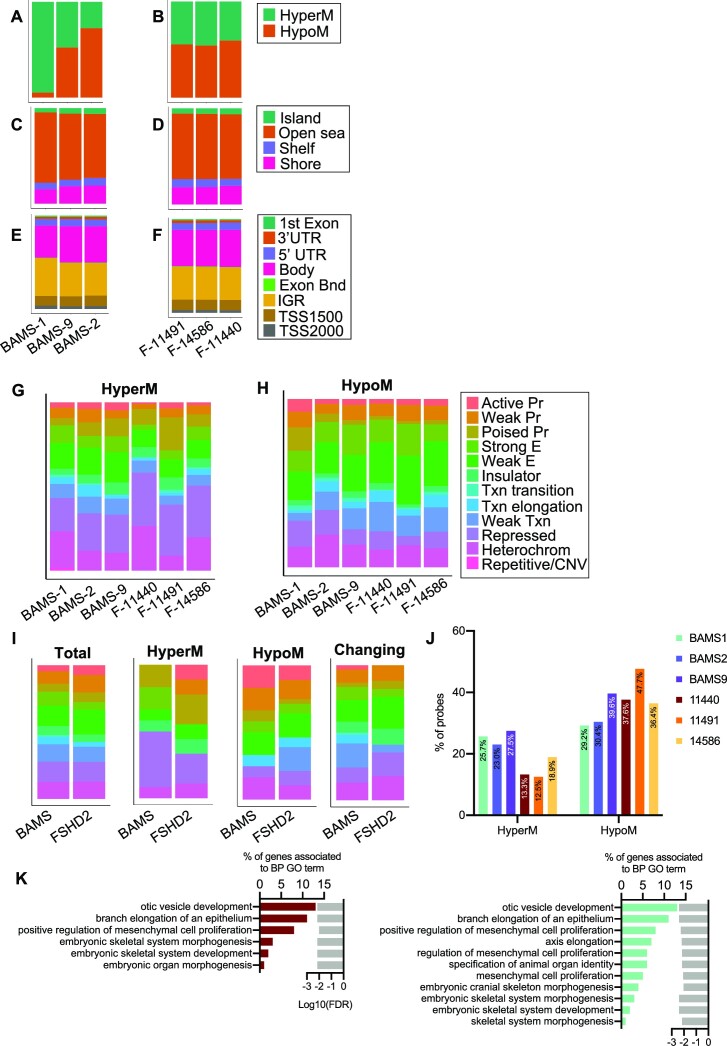
Figure 1.
Profiling of DNAme in SMCHD1-deficient cells. One hundred percent stacked bar graphs of the distribution of hypomethylated and hypermethylated probes in individual BAMS samples (A) or individual FSHD2 samples (B). One hundred percent stacked bar graphs for DMPs by CpG content relative to CpG islands, shores (2 kb flanking CpG islands), shelves (2 kb extending from shores) or open seas (isolated CpGs in the rest of the genome) in individual BAMS samples (C) or individual FSHD2 samples (D). One hundred percent stacked bar graphs for DMPs by features corresponding to genes’ first exon, 3′ UTR, 5′ UTR, gene bodies, exon boundaries, internal genomic regions and probes located 1500 bp from transcription start sites (TSS1500) or 2000 bp from transcription start sites (TSS200) in individual BAMS samples (E) or individual FSHD2 samples (F). DMRs with an FDR-adjusted P-value <0.05 were analyzed for ChromHMM features using NHEK cell annotations and plotted using the ggplot2 (v3.3.3) R package. One hundred percent stacked bar graphs for hypermethylated (G) or hypomethylated (H) probes are presented. (I) Left to right: bar plots of DMRs shared between patients when compared to controls (total), shared hypermethylated DMRs (HyperM), shared hypomethylated DMRs (HypoM) and DMRs with a different methylation profile between patients (changing). (J) Graph displaying the percentage of hypermethylated or hypomethylated probes at enhancers in the different BAMS and FSHD2 samples. (K) BPs overrepresented for BAMS (cyan, right) or FSHD2 (red, left) DMRs. Bar plots on the left represent the percentage of genes and associated with GO terms listed in the right column. Light gray bars on the right represent the enrichment score (log10FDR) for each GO term.