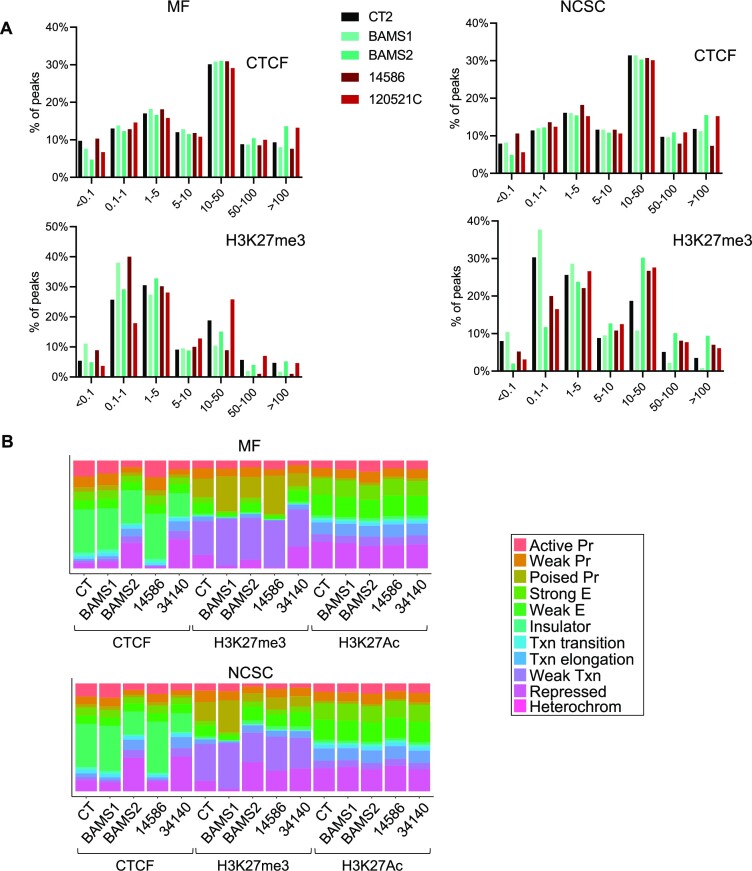
Figure 3.
Chromatin profiling of hiPSC-derived MFs and NCSCs from SMCHD1-deficient patients. (A) Distribution of distances from TSSs for peaks enriched in CTCF or H3K27me3 in controls, BAMS or FSHD2 MFs or NCSCs. Peaks’ distribution relative to TSSs was assessed using the chipenrich (v2.14.0) R package. (B) Distribution of peaks enriched in CTCF, H3K27me3 or H3K27Ac relative to chromatin features determined using the ChromHMM track in MFs or NCSCs derived from control, BAMS or FSHD hiPSCs. Peaks with at least a q-value <0.05 in one replicate were analyzed for ChromHMM features using HSMM cell annotations and represented as bar plots using the R ggplot2 (v3.3.3) package.