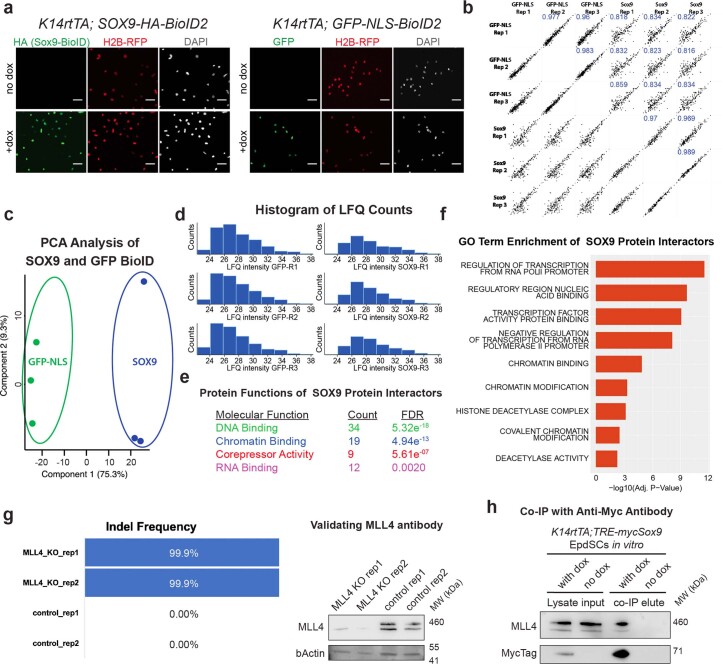
Extended Data Fig. 5. BioID experiments and identification of SOX9 co-factors.
a, Immunofluorescence validation of Krt14rtTA primary keratinocytes transduced with TRE-HA-SOX9-BioID2; H2B-RFP (left) or TRE-GFP-NLS-BioID2; H2B-RFP (right). Transgenes were induced with doxycycline and immunolabeled for HA (left) or GFP (right). All scale bars are 50μm. b, Correlation plots of Label Free Quantification (LFQ) values identified by BioID experiments across replicates and samples. SOX9-BioID and GFP-NLS-BioID samples share stronger correlations between replicates than each other. Blue number represents r2 value. c, PCA analysis of GFP and SOX9 BioID protein interactors demonstrate sample-specific clusters. d, Histogram of LFQ intensity values for the GFP (left) and SOX (right) BioID replicates. e, Molecular function enrichment of proteins specifically interacting with SOX9. f, Gene ontology enrichment of proteins specific to SOX9 (binomial is used to calculate the significance). g, Two Mll4 null keratinocyte cell lines (Krt14rtTA; TRE-mycSox9) were generated with CRIPSR/Cas9 with 99.9% indel frequency. These lines validated the efficacy of the MLL4 antibody by immunoblot, which detected a ~500 kDa protein in the control but not the knockout (KO) cell lines. MW, molecular weight. h, Immunoblot showing that the ~500 kDa protein, identified in (g) as MLL4, is pulled down with mycSOX9 in an anti-myc tag antibody immunoprecipitation. See also in associated source data.