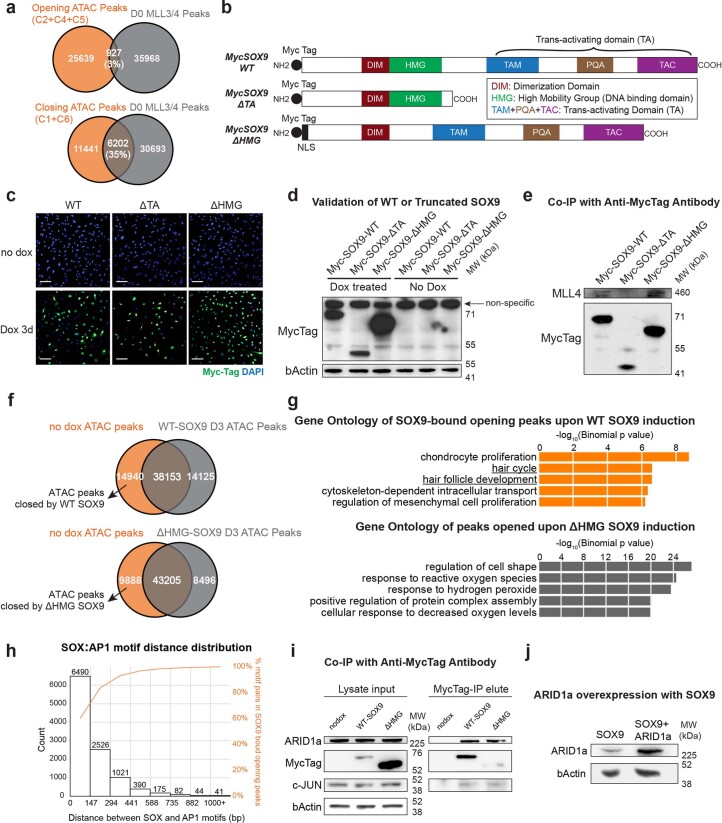
Extended Data Fig. 7. Truncated SOX9 displays impaired DNA binding or co-factor recruitment.
a, (top) Venn diagram reveals almost no overlap between ATAC peaks (C2,C4,C5) that mostly open between W2-W12 and D0 MLL3/4 Cut-and-Run peaks. (bottom) Venn diagram shows substantial overlap between ATAC peaks (C1,C6) that mostly close between W1-W2 and D0 MLL3/4 Cut-and-Run peaks. b, Schematic illustrating constructs engineered to express WT SOX9 and two variants of SOX9. c, Immunofluorescence reveals similar intensities of WT and mutant SOX9 in induced EpdSCs. Scale bars, 100μm. d, Immunoblot validating the sizes of different versions of SOX9. MW, molecular weight. e, Immunoblot showing that the transactivating (TA) domain of SOX9 is sufficient in binding MLL4. f, Venn diagrams show that the peak sets closed by WT and ΔHMG-SOX9 are comparable in size. g, (top) Top 5 biological process gene ontologies of genes associated with SOX9-bound opening peaks upon WT SOX9 induction in vitro from GREAT. (bottom) Top 5 biological process gene ontologies of genes associated with peaks opened upon ΔHMG SOX9 induction in vitro from GREAT. Binomial is used to calculate the significance. h, Distribution of the distance between a SOX motif and its closest AP1 motif in the SOX9 bound opening peaks. Note that the x-axis is binned by multiplies of one nucleosome size (147bp). The cumulation plot of the distribution is shown in orange on the secondary y-axis. i, Immunoblot showing that both WT-SOX9 and ΔHMG-SOX9 are capable of binding c-JUN and ARID1a. j, Immunoblot validating ARID1a can be induced 3x higher in the SOX9 expressing keratinocytes. For full blots, see associated source data.